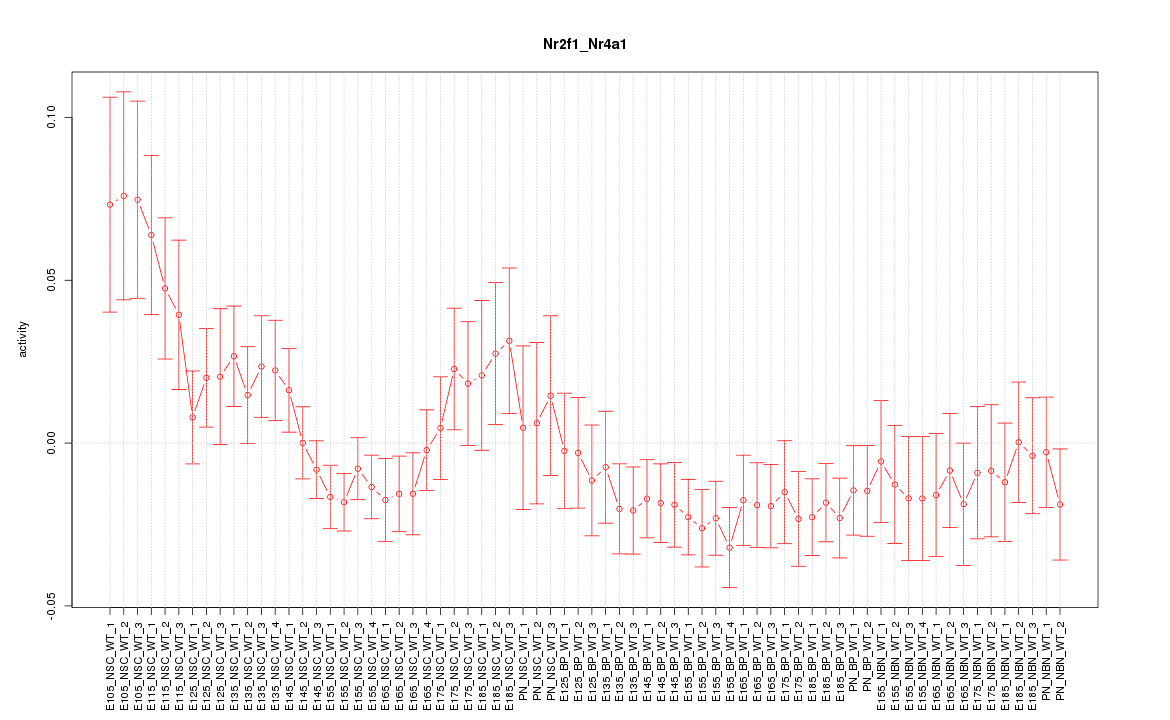
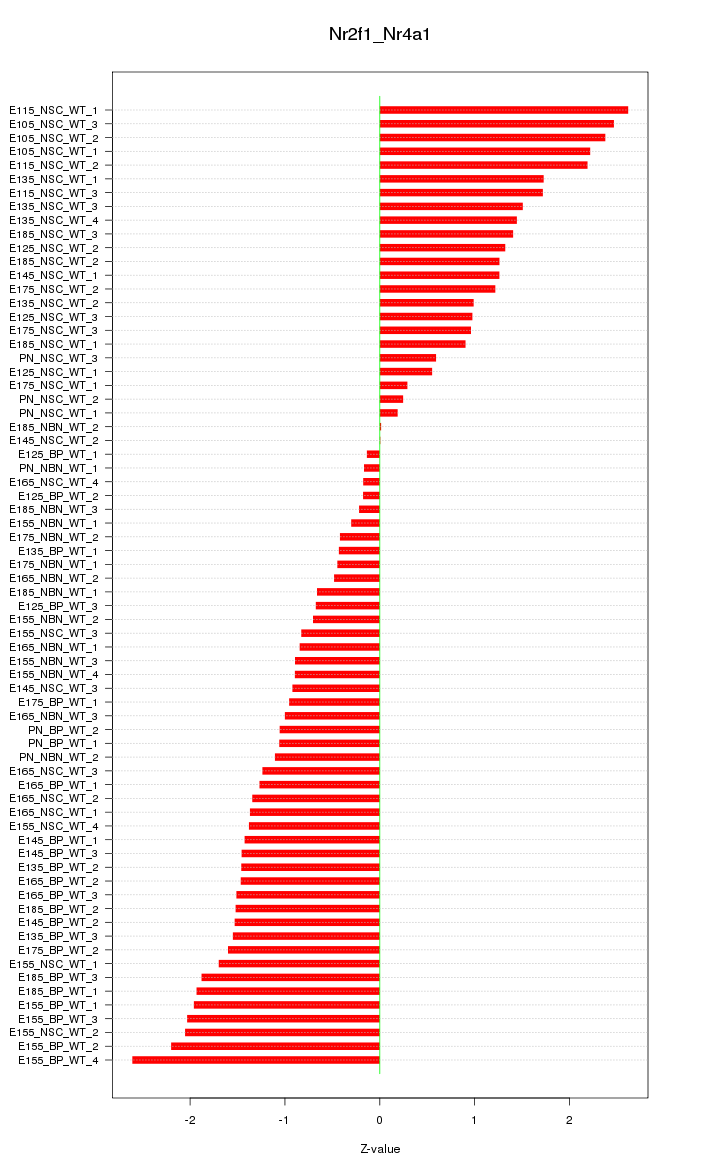
Motif ID: Nr2f1_Nr4a1
Z-value: 1.343
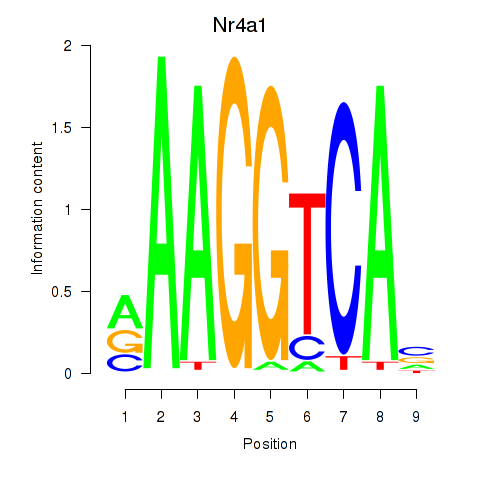
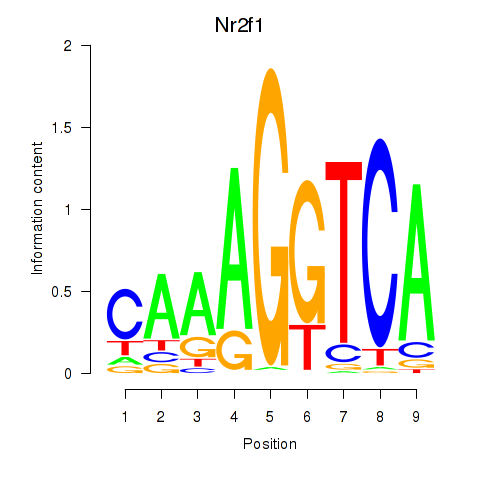
Transcription factors associated with Nr2f1_Nr4a1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Nr2f1 | ENSMUSG00000069171.7 | Nr2f1 |
| Nr4a1 | ENSMUSG00000023034.6 | Nr4a1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr2f1 | mm10_v2_chr13_-_78199757_78199855 | -0.36 | 2.3e-03 | Click! |
| Nr4a1 | mm10_v2_chr15_+_101266839_101266859 | -0.11 | 3.5e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.5 | 16.5 | GO:0042822 | vitamin B6 metabolic process(GO:0042816) pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 5.3 | 16.0 | GO:0014877 | response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) membrane depolarization during SA node cell action potential(GO:0086046) |
| 4.9 | 19.7 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 4.4 | 22.1 | GO:0014719 | skeletal muscle satellite cell activation(GO:0014719) |
| 4.1 | 16.2 | GO:2000974 | negative regulation of pro-B cell differentiation(GO:2000974) |
| 3.9 | 11.8 | GO:0046544 | regulation of natural killer cell proliferation(GO:0032817) positive regulation of natural killer cell proliferation(GO:0032819) development of secondary male sexual characteristics(GO:0046544) |
| 3.1 | 12.4 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 2.9 | 8.8 | GO:1903795 | regulation of inorganic anion transmembrane transport(GO:1903795) |
| 2.9 | 8.7 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 2.9 | 8.6 | GO:0072076 | nephrogenic mesenchyme development(GO:0072076) |
| 2.6 | 7.7 | GO:0035672 | regulation of cellular pH reduction(GO:0032847) oligopeptide transmembrane transport(GO:0035672) |
| 2.4 | 7.3 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 2.4 | 7.3 | GO:0097461 | ferric iron import(GO:0033216) ferric iron import into cell(GO:0097461) |
| 2.4 | 14.7 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 2.4 | 7.3 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 2.3 | 11.5 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 2.2 | 6.5 | GO:0003360 | brainstem development(GO:0003360) |
| 2.1 | 12.9 | GO:0001980 | regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 2.1 | 6.3 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 2.1 | 8.4 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 2.0 | 10.2 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 1.8 | 9.2 | GO:0021905 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) |
| 1.7 | 5.2 | GO:2000983 | regulation of ATP citrate synthase activity(GO:2000983) negative regulation of ATP citrate synthase activity(GO:2000984) |
| 1.7 | 5.1 | GO:0032493 | response to bacterial lipoprotein(GO:0032493) |
| 1.7 | 5.0 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 1.7 | 8.3 | GO:0046133 | pyrimidine ribonucleoside catabolic process(GO:0046133) |
| 1.6 | 4.8 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 1.6 | 11.0 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 1.6 | 4.7 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 1.5 | 4.6 | GO:0042560 | 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 1.5 | 4.6 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 1.5 | 6.0 | GO:0006867 | asparagine transport(GO:0006867) glutamine transport(GO:0006868) |
| 1.5 | 4.4 | GO:0000087 | mitotic M phase(GO:0000087) |
| 1.4 | 4.3 | GO:0015889 | cobalamin transport(GO:0015889) |
| 1.4 | 2.8 | GO:1990705 | cholangiocyte proliferation(GO:1990705) |
| 1.4 | 6.9 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 1.4 | 1.4 | GO:0046135 | pyrimidine nucleoside catabolic process(GO:0046135) |
| 1.3 | 12.1 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 1.3 | 6.5 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 1.3 | 3.9 | GO:0051695 | actin filament uncapping(GO:0051695) |
| 1.2 | 6.1 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 1.2 | 3.6 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 1.2 | 3.6 | GO:0060437 | lung growth(GO:0060437) |
| 1.2 | 4.8 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 1.2 | 5.8 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 1.1 | 5.7 | GO:0090234 | cellular response to testosterone stimulus(GO:0071394) regulation of kinetochore assembly(GO:0090234) |
| 1.1 | 6.6 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 1.1 | 3.2 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 1.0 | 3.1 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 1.0 | 4.1 | GO:0033762 | response to glucagon(GO:0033762) |
| 1.0 | 5.0 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 1.0 | 4.0 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 1.0 | 4.9 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 1.0 | 8.8 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 1.0 | 3.8 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.9 | 5.7 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.9 | 2.8 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.9 | 3.6 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.9 | 2.7 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.9 | 3.6 | GO:0060745 | mammary gland branching involved in pregnancy(GO:0060745) |
| 0.9 | 4.4 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.9 | 2.7 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.9 | 4.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.9 | 3.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.9 | 9.4 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.8 | 1.7 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.8 | 4.8 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.8 | 4.6 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.8 | 2.3 | GO:0021546 | rhombomere development(GO:0021546) |
| 0.7 | 5.7 | GO:0002681 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.7 | 2.9 | GO:0002741 | positive regulation of cytokine secretion involved in immune response(GO:0002741) |
| 0.7 | 2.1 | GO:0034035 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) |
| 0.7 | 2.1 | GO:0009174 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.7 | 2.1 | GO:0090202 | regulation of primitive erythrocyte differentiation(GO:0010725) transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.7 | 4.2 | GO:0098700 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.7 | 2.8 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.7 | 4.0 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.7 | 2.0 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.7 | 4.0 | GO:0035989 | tendon development(GO:0035989) |
| 0.7 | 18.3 | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.6 | 18.2 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.6 | 3.9 | GO:0042535 | positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) macropinocytosis(GO:0044351) |
| 0.6 | 1.9 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.6 | 10.3 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.6 | 22.4 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.6 | 4.3 | GO:1904667 | negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.6 | 4.7 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.6 | 1.7 | GO:0003100 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) |
| 0.6 | 3.9 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.6 | 1.1 | GO:2000157 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.5 | 2.7 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.5 | 4.3 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.5 | 5.4 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.5 | 2.1 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.5 | 4.8 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.5 | 1.6 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.5 | 14.8 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.5 | 4.6 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.5 | 2.6 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.5 | 3.6 | GO:0046459 | short-chain fatty acid metabolic process(GO:0046459) |
| 0.5 | 2.0 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.5 | 1.5 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.5 | 1.5 | GO:0090274 | positive regulation of pancreatic juice secretion(GO:0090187) positive regulation of somatostatin secretion(GO:0090274) |
| 0.5 | 3.9 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.5 | 1.5 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.5 | 3.4 | GO:0072257 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.5 | 1.9 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.5 | 4.7 | GO:0046060 | dATP metabolic process(GO:0046060) |
| 0.5 | 0.9 | GO:2000338 | chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 0.5 | 0.9 | GO:0099624 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) atrial cardiac muscle cell membrane repolarization(GO:0099624) |
| 0.5 | 2.3 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.4 | 3.6 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.4 | 6.7 | GO:0097154 | GABAergic neuron differentiation(GO:0097154) |
| 0.4 | 5.8 | GO:0006260 | DNA replication(GO:0006260) |
| 0.4 | 1.8 | GO:0006842 | tricarboxylic acid transport(GO:0006842) succinate transport(GO:0015744) citrate transport(GO:0015746) |
| 0.4 | 1.3 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.4 | 15.4 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.4 | 3.5 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.4 | 6.9 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.4 | 1.3 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.4 | 1.3 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.4 | 4.3 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.4 | 2.1 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) negative regulation of neutrophil activation(GO:1902564) |
| 0.4 | 2.9 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.4 | 1.2 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) alveolar secondary septum development(GO:0061144) |
| 0.4 | 0.4 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.4 | 2.1 | GO:0045918 | negative regulation of cytolysis(GO:0045918) |
| 0.4 | 2.9 | GO:0021780 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.4 | 2.4 | GO:0043586 | tongue development(GO:0043586) |
| 0.4 | 5.6 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.4 | 1.2 | GO:0019043 | establishment of viral latency(GO:0019043) |
| 0.4 | 19.1 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.4 | 2.0 | GO:1900085 | negative regulation of pinocytosis(GO:0048550) negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) |
| 0.4 | 0.8 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.4 | 1.5 | GO:0050904 | diapedesis(GO:0050904) |
| 0.4 | 4.5 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.4 | 0.7 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.4 | 1.8 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.4 | 2.5 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.4 | 2.5 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.4 | 2.8 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.4 | 1.8 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.4 | 0.7 | GO:0048162 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.3 | 1.0 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.3 | 2.8 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.3 | 1.4 | GO:2000383 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.3 | 1.3 | GO:0010694 | positive regulation of alkaline phosphatase activity(GO:0010694) |
| 0.3 | 2.5 | GO:0036376 | sodium ion export from cell(GO:0036376) |
| 0.3 | 0.6 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.3 | 0.3 | GO:0044830 | modulation by host of viral RNA genome replication(GO:0044830) |
| 0.3 | 1.2 | GO:0015786 | UDP-glucose transport(GO:0015786) |
| 0.3 | 3.3 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.3 | 3.9 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.3 | 5.1 | GO:0002052 | positive regulation of neuroblast proliferation(GO:0002052) |
| 0.3 | 1.5 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.3 | 0.3 | GO:0036115 | fatty-acyl-CoA catabolic process(GO:0036115) |
| 0.3 | 3.2 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) sphingoid biosynthetic process(GO:0046520) |
| 0.3 | 3.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.3 | 0.6 | GO:2000508 | oligodendrocyte apoptotic process(GO:0097252) regulation of dendritic cell chemotaxis(GO:2000508) positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.3 | 1.7 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.3 | 3.2 | GO:0014883 | regulation of skeletal muscle adaptation(GO:0014733) transition between fast and slow fiber(GO:0014883) skeletal muscle adaptation(GO:0043501) |
| 0.3 | 0.9 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.3 | 0.6 | GO:0061419 | positive regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061419) |
| 0.3 | 2.3 | GO:1903624 | regulation of apoptotic DNA fragmentation(GO:1902510) regulation of DNA catabolic process(GO:1903624) |
| 0.3 | 4.7 | GO:0010883 | regulation of lipid storage(GO:0010883) |
| 0.3 | 3.9 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.3 | 0.8 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.3 | 1.4 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.3 | 1.1 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.3 | 4.1 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.3 | 1.6 | GO:1903755 | regulation of SUMO transferase activity(GO:1903182) positive regulation of SUMO transferase activity(GO:1903755) |
| 0.3 | 4.0 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.3 | 6.4 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.3 | 0.8 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.3 | 3.4 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.2 | 4.9 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.2 | 1.2 | GO:0045840 | positive regulation of mitotic nuclear division(GO:0045840) |
| 0.2 | 0.7 | GO:0030300 | regulation of intestinal cholesterol absorption(GO:0030300) |
| 0.2 | 1.7 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.2 | 3.0 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.2 | 1.4 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.2 | 3.9 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.2 | 6.0 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.2 | 3.9 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.2 | 1.1 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.2 | 1.1 | GO:0060028 | non-canonical Wnt signaling pathway via MAPK cascade(GO:0038030) non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) convergent extension involved in axis elongation(GO:0060028) |
| 0.2 | 0.7 | GO:0001193 | maintenance of transcriptional fidelity during DNA-templated transcription elongation(GO:0001192) maintenance of transcriptional fidelity during DNA-templated transcription elongation from RNA polymerase II promoter(GO:0001193) |
| 0.2 | 0.6 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.2 | 3.9 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.2 | 14.1 | GO:0006096 | glycolytic process(GO:0006096) ATP generation from ADP(GO:0006757) |
| 0.2 | 0.6 | GO:0031627 | telomeric loop formation(GO:0031627) |
| 0.2 | 2.1 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.2 | 1.0 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.2 | 0.8 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.2 | 6.6 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.2 | 7.2 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.2 | 1.0 | GO:0060576 | O-glycan processing(GO:0016266) intestinal epithelial cell development(GO:0060576) |
| 0.2 | 2.7 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.2 | 1.2 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.2 | 0.8 | GO:0061525 | hindgut development(GO:0061525) |
| 0.2 | 2.0 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.2 | 5.6 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.2 | 2.4 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.2 | 14.8 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.2 | 7.6 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.2 | 6.3 | GO:0030865 | cortical cytoskeleton organization(GO:0030865) |
| 0.2 | 2.4 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.2 | 2.0 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.2 | 2.8 | GO:0031065 | positive regulation of histone deacetylation(GO:0031065) |
| 0.2 | 4.6 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.2 | 0.7 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.2 | 1.3 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.2 | 3.0 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.2 | 0.3 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.2 | 1.1 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.2 | 0.9 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.2 | 2.6 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.2 | 1.6 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.2 | 1.2 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.2 | 0.9 | GO:0046929 | negative regulation of neurotransmitter secretion(GO:0046929) |
| 0.2 | 0.8 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.2 | 0.5 | GO:0000467 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.1 | 2.8 | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.1 | 1.6 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.1 | 4.8 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.1 | 9.0 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.1 | 2.0 | GO:0033866 | coenzyme A biosynthetic process(GO:0015937) nucleoside bisphosphate biosynthetic process(GO:0033866) ribonucleoside bisphosphate biosynthetic process(GO:0034030) purine nucleoside bisphosphate biosynthetic process(GO:0034033) |
| 0.1 | 1.0 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.1 | 2.2 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.1 | 0.6 | GO:0065001 | negative regulation of histone H3-K36 methylation(GO:0000415) specification of axis polarity(GO:0065001) |
| 0.1 | 1.7 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.1 | 2.1 | GO:0006402 | mRNA catabolic process(GO:0006402) |
| 0.1 | 1.0 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 1.0 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 | 2.1 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 0.3 | GO:0019530 | taurine metabolic process(GO:0019530) |
| 0.1 | 2.5 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.1 | 0.9 | GO:0003091 | renal water homeostasis(GO:0003091) |
| 0.1 | 7.8 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.1 | 0.5 | GO:2000973 | regulation of pro-B cell differentiation(GO:2000973) |
| 0.1 | 0.9 | GO:0015677 | copper ion import(GO:0015677) |
| 0.1 | 0.9 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.1 | 0.4 | GO:0060440 | trachea formation(GO:0060440) regulation of Golgi inheritance(GO:0090170) |
| 0.1 | 0.8 | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.1 | 1.7 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 3.7 | GO:0031050 | dsRNA fragmentation(GO:0031050) production of miRNAs involved in gene silencing by miRNA(GO:0035196) production of small RNA involved in gene silencing by RNA(GO:0070918) |
| 0.1 | 1.4 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.1 | 0.6 | GO:0060766 | negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.1 | 1.5 | GO:0010666 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.1 | 1.0 | GO:0035584 | calcium-mediated signaling using intracellular calcium source(GO:0035584) |
| 0.1 | 14.7 | GO:0006006 | glucose metabolic process(GO:0006006) |
| 0.1 | 2.8 | GO:0031111 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.1 | 0.8 | GO:0090344 | negative regulation of cell aging(GO:0090344) |
| 0.1 | 4.9 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.1 | 3.1 | GO:0042026 | protein refolding(GO:0042026) |
| 0.1 | 1.4 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.1 | 1.5 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.1 | 0.4 | GO:0045346 | regulation of MHC class II biosynthetic process(GO:0045346) negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.1 | 1.7 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.1 | 1.1 | GO:0035814 | negative regulation of renal sodium excretion(GO:0035814) |
| 0.1 | 2.3 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 0.7 | GO:0061684 | chaperone-mediated autophagy(GO:0061684) |
| 0.1 | 1.3 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 3.2 | GO:0006220 | pyrimidine nucleotide metabolic process(GO:0006220) |
| 0.1 | 2.2 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 1.2 | GO:0006337 | nucleosome disassembly(GO:0006337) protein-DNA complex disassembly(GO:0032986) |
| 0.1 | 1.8 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.1 | 0.3 | GO:0006706 | steroid catabolic process(GO:0006706) |
| 0.1 | 1.4 | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) |
| 0.1 | 1.8 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 0.7 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.1 | 2.1 | GO:0035886 | vascular smooth muscle cell differentiation(GO:0035886) |
| 0.1 | 1.5 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.1 | 0.3 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 1.5 | GO:0007614 | short-term memory(GO:0007614) |
| 0.1 | 4.3 | GO:0016055 | Wnt signaling pathway(GO:0016055) cell-cell signaling by wnt(GO:0198738) |
| 0.1 | 4.2 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.1 | 0.5 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 1.1 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.1 | 3.7 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.1 | 0.4 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.1 | 1.0 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.1 | 0.3 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.1 | 0.2 | GO:0019478 | D-amino acid catabolic process(GO:0019478) |
| 0.1 | 1.2 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 0.6 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.1 | 0.4 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.1 | 1.2 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.1 | 0.5 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 6.0 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 1.5 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.1 | 0.2 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.1 | 4.5 | GO:0001892 | embryonic placenta development(GO:0001892) |
| 0.1 | 2.2 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.1 | 1.7 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.1 | 0.3 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.1 | 1.1 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.1 | 1.0 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 0.3 | GO:1903887 | motile primary cilium assembly(GO:1903887) |
| 0.1 | 0.4 | GO:0007143 | female meiotic division(GO:0007143) |
| 0.1 | 1.6 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.1 | 5.6 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.1 | 0.9 | GO:0035194 | posttranscriptional gene silencing(GO:0016441) posttranscriptional gene silencing by RNA(GO:0035194) gene silencing by miRNA(GO:0035195) |
| 0.1 | 4.2 | GO:0070374 | positive regulation of ERK1 and ERK2 cascade(GO:0070374) |
| 0.1 | 0.4 | GO:1903504 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.1 | 1.1 | GO:0035036 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) |
| 0.1 | 1.4 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.4 | GO:0007249 | I-kappaB kinase/NF-kappaB signaling(GO:0007249) |
| 0.1 | 0.2 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.1 | 0.1 | GO:0034140 | negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) |
| 0.1 | 0.5 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.0 | 1.4 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 4.4 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 0.1 | GO:0042373 | fat-soluble vitamin biosynthetic process(GO:0042362) vitamin K metabolic process(GO:0042373) |
| 0.0 | 0.7 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 1.6 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.9 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.3 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.6 | GO:0097006 | regulation of plasma lipoprotein particle levels(GO:0097006) |
| 0.0 | 0.2 | GO:0072429 | negative regulation of chromatin binding(GO:0035562) response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 | 0.9 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.0 | 1.7 | GO:0048144 | fibroblast proliferation(GO:0048144) |
| 0.0 | 1.5 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.2 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.0 | 1.0 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.8 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 1.0 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 0.2 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.2 | GO:0002385 | organ or tissue specific immune response(GO:0002251) mucosal immune response(GO:0002385) |
| 0.0 | 0.9 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 1.0 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 3.2 | GO:0006626 | protein targeting to mitochondrion(GO:0006626) |
| 0.0 | 2.0 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.5 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 2.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.1 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 4.8 | GO:0007059 | chromosome segregation(GO:0007059) |
| 0.0 | 0.1 | GO:0051189 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.1 | GO:1904666 | regulation of ubiquitin protein ligase activity(GO:1904666) |
| 0.0 | 0.9 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.2 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.7 | GO:0048661 | positive regulation of smooth muscle cell proliferation(GO:0048661) |
| 0.0 | 0.8 | GO:0043507 | positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 0.5 | GO:0021987 | cerebral cortex development(GO:0021987) |
| 0.0 | 2.3 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.2 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.5 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.9 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.2 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.3 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 1.3 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 0.4 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.7 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 1.4 | GO:0006364 | rRNA processing(GO:0006364) rRNA metabolic process(GO:0016072) |
| 0.0 | 0.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.7 | GO:0000725 | double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 0.0 | 0.5 | GO:0061515 | myeloid cell development(GO:0061515) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.9 | 14.7 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 3.6 | 18.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 2.6 | 7.8 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 2.6 | 12.9 | GO:0005861 | troponin complex(GO:0005861) |
| 2.2 | 6.5 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 1.6 | 4.7 | GO:0031983 | vesicle lumen(GO:0031983) |
| 1.3 | 8.1 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 1.3 | 5.2 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 1.3 | 8.8 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 1.2 | 10.0 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 1.2 | 8.4 | GO:0097452 | GAIT complex(GO:0097452) |
| 1.2 | 3.6 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 1.1 | 12.9 | GO:0000124 | SAGA complex(GO:0000124) |
| 1.0 | 7.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.9 | 4.7 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 0.8 | 2.5 | GO:0036125 | mitochondrial fatty acid beta-oxidation multienzyme complex(GO:0016507) fatty acid beta-oxidation multienzyme complex(GO:0036125) |
| 0.8 | 13.0 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.8 | 3.0 | GO:0045251 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.8 | 6.8 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.7 | 11.5 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.7 | 8.5 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.7 | 4.0 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.6 | 14.8 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.6 | 6.5 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.6 | 5.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.6 | 2.2 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.6 | 4.4 | GO:0031415 | NatA complex(GO:0031415) |
| 0.5 | 8.2 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.5 | 2.6 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.5 | 5.7 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.5 | 1.5 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.5 | 4.9 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.5 | 2.9 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.5 | 1.4 | GO:0031251 | PAN complex(GO:0031251) |
| 0.4 | 4.0 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.4 | 1.3 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.4 | 1.3 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.4 | 2.6 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.4 | 1.3 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.4 | 7.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.4 | 19.2 | GO:0098803 | respiratory chain complex(GO:0098803) |
| 0.4 | 4.0 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.4 | 5.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.4 | 2.0 | GO:0034715 | U7 snRNP(GO:0005683) pICln-Sm protein complex(GO:0034715) |
| 0.4 | 1.6 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.4 | 1.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.4 | 3.0 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.4 | 4.1 | GO:0030057 | desmosome(GO:0030057) |
| 0.4 | 6.0 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.4 | 3.3 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.4 | 3.7 | GO:0046930 | pore complex(GO:0046930) |
| 0.4 | 7.4 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.4 | 3.6 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.3 | 5.0 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.3 | 0.7 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.3 | 3.4 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.3 | 0.9 | GO:0070449 | elongin complex(GO:0070449) |
| 0.3 | 4.0 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.3 | 22.2 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.3 | 1.4 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.3 | 1.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.3 | 6.1 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.3 | 1.3 | GO:0031523 | Myb complex(GO:0031523) |
| 0.2 | 1.2 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.2 | 1.0 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.2 | 1.4 | GO:0032009 | early phagosome(GO:0032009) |
| 0.2 | 0.7 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.2 | 1.4 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.2 | 0.9 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.2 | 6.7 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.2 | 12.8 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.2 | 11.8 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.2 | 0.6 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.2 | 3.1 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.2 | 3.6 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.2 | 0.8 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.2 | 2.9 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.2 | 0.4 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.2 | 1.1 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.2 | 4.0 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.2 | 1.1 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.2 | 1.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.2 | 2.7 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.2 | 6.0 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.2 | 1.0 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.2 | 1.7 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.2 | 1.6 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.2 | 3.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.2 | 2.4 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.2 | 0.6 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.1 | 5.8 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 1.0 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 12.4 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 25.6 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 0.7 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 3.2 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 0.6 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.1 | 1.5 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 1.4 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.1 | 6.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 4.7 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 0.4 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.1 | 9.7 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 4.7 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 44.1 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 1.5 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 6.0 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 1.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 23.2 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.1 | 0.4 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 2.8 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 0.7 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 2.2 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.1 | 1.3 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 0.9 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 2.5 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 1.6 | GO:0030120 | vesicle coat(GO:0030120) |
| 0.1 | 20.7 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 0.8 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 1.0 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 1.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 1.2 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.1 | 1.0 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 1.2 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.1 | 4.0 | GO:0015934 | large ribosomal subunit(GO:0015934) |
| 0.1 | 4.4 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 0.5 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 5.0 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 0.3 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 0.8 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 1.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 0.6 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.1 | 1.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 0.3 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 4.2 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.1 | 2.6 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 2.1 | GO:0044391 | ribosomal subunit(GO:0044391) |
| 0.0 | 7.6 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 1.0 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.3 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 1.5 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 1.1 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.5 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.3 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 1.1 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 18.6 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.4 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.7 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 1.2 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.1 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.9 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.5 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.2 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 2.3 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.6 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.1 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 1.1 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 2.0 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.4 | GO:0030118 | clathrin coat(GO:0030118) |
| 0.0 | 0.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.3 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 1.9 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.5 | 16.5 | GO:0031403 | lithium ion binding(GO:0031403) |
| 4.3 | 12.9 | GO:0030172 | troponin C binding(GO:0030172) |
| 4.0 | 12.1 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 3.9 | 19.7 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 3.5 | 17.7 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 2.8 | 8.4 | GO:0035605 | peptidyl-cysteine S-nitrosylase activity(GO:0035605) |
| 2.6 | 28.6 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 2.5 | 10.2 | GO:0038049 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 2.4 | 7.3 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 2.0 | 18.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 2.0 | 7.8 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 1.9 | 7.7 | GO:0004064 | arylesterase activity(GO:0004064) |
| 1.9 | 5.6 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 1.9 | 13.0 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 1.7 | 6.8 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 1.6 | 6.5 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 1.6 | 4.7 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 1.5 | 4.6 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 1.5 | 4.6 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) bisphosphoglycerate 2-phosphatase activity(GO:0004083) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 1.5 | 6.1 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 1.5 | 6.0 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 1.5 | 10.2 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 1.4 | 2.9 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 1.3 | 4.0 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 1.3 | 5.2 | GO:1990254 | keratin filament binding(GO:1990254) |
| 1.3 | 7.6 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 1.3 | 7.6 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 1.3 | 3.8 | GO:0048045 | 4-hydroxybenzoate octaprenyltransferase activity(GO:0008412) protoheme IX farnesyltransferase activity(GO:0008495) (S)-2,3-di-O-geranylgeranylglyceryl phosphate synthase activity(GO:0043888) cadaverine aminopropyltransferase activity(GO:0043918) agmatine aminopropyltransferase activity(GO:0043919) 1,4-dihydroxy-2-naphthoate octaprenyltransferase activity(GO:0046428) trans-pentaprenyltranstransferase activity(GO:0048045) ATP dimethylallyltransferase activity(GO:0052622) ADP dimethylallyltransferase activity(GO:0052623) |
| 1.2 | 6.0 | GO:0015186 | L-asparagine transmembrane transporter activity(GO:0015182) L-glutamine transmembrane transporter activity(GO:0015186) |
| 1.2 | 3.6 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 1.2 | 5.8 | GO:0004359 | glutaminase activity(GO:0004359) |
| 1.1 | 6.8 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 1.1 | 10.2 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 1.0 | 4.2 | GO:0036468 | aromatic-L-amino-acid decarboxylase activity(GO:0004058) L-dopa decarboxylase activity(GO:0036468) |
| 1.0 | 12.4 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 1.0 | 3.1 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 1.0 | 4.0 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 1.0 | 4.0 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 1.0 | 9.9 | GO:0008556 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) |
| 1.0 | 5.8 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.9 | 2.8 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.9 | 2.7 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.9 | 10.8 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.9 | 5.1 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.8 | 9.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.8 | 11.0 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.8 | 6.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.7 | 3.6 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.7 | 2.1 | GO:0004658 | propionyl-CoA carboxylase activity(GO:0004658) |
| 0.7 | 3.9 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.6 | 3.9 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.6 | 6.4 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.6 | 2.5 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.6 | 1.9 | GO:0008311 | double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.6 | 4.3 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.6 | 4.9 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.6 | 1.8 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.6 | 1.7 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.6 | 1.7 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.6 | 5.1 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.5 | 4.8 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.5 | 3.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.5 | 1.6 | GO:0070890 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.5 | 5.3 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.5 | 5.3 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.5 | 11.5 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.5 | 3.6 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.5 | 2.5 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.5 | 2.5 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.5 | 5.7 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.5 | 12.7 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.5 | 2.3 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.5 | 2.3 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.5 | 3.7 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.4 | 9.7 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.4 | 17.2 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.4 | 3.0 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.4 | 2.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.4 | 2.5 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.4 | 1.2 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.4 | 3.6 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.4 | 2.0 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.4 | 4.7 | GO:0005167 | neurotrophin TRK receptor binding(GO:0005167) |
| 0.4 | 13.0 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.4 | 1.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) |
| 0.4 | 1.9 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.4 | 2.6 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.4 | 7.5 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.4 | 1.5 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.4 | 5.5 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.4 | 7.2 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.4 | 2.1 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.4 | 1.8 | GO:0008808 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) |
| 0.3 | 4.4 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.3 | 1.4 | GO:0002054 | nucleobase binding(GO:0002054) |
| 0.3 | 6.9 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.3 | 1.3 | GO:0098634 | protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.3 | 5.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.3 | 1.0 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.3 | 5.5 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.3 | 1.3 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.3 | 0.9 | GO:0004008 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.3 | 0.9 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.3 | 4.9 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.3 | 2.8 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.3 | 7.1 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.3 | 0.8 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.3 | 1.6 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.3 | 10.4 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.3 | 1.3 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.3 | 1.5 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.2 | 9.0 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) |
| 0.2 | 3.0 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.2 | 2.4 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.2 | 14.9 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.2 | 1.0 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.2 | 2.4 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.2 | 1.6 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.2 | 6.3 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.2 | 0.9 | GO:0070976 | TIR domain binding(GO:0070976) |
| 0.2 | 6.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.2 | 5.2 | GO:0016667 | oxidoreductase activity, acting on a sulfur group of donors(GO:0016667) |
| 0.2 | 2.7 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.2 | 1.1 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.2 | 0.6 | GO:0071862 | protein phosphatase type 1 activator activity(GO:0071862) |
| 0.2 | 1.1 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.2 | 3.9 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.2 | 0.8 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.2 | 0.6 | GO:0002134 | UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.2 | 1.4 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.2 | 0.6 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.2 | 2.3 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.2 | 3.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.2 | 3.4 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.2 | 0.7 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.2 | 1.1 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.2 | 8.3 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.2 | 0.9 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.2 | 1.6 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.2 | 2.8 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.2 | 0.5 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.2 | 3.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.2 | 6.1 | GO:0033549 | MAP kinase phosphatase activity(GO:0033549) |
| 0.2 | 3.8 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.2 | 10.6 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.2 | 2.6 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 21.7 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.1 | 20.2 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 0.6 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.1 | 1.4 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 5.7 | GO:0052771 | coenzyme F390-A hydrolase activity(GO:0052770) coenzyme F390-G hydrolase activity(GO:0052771) |
| 0.1 | 0.5 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.1 | 2.1 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 4.8 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.1 | 2.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 1.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.7 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 7.4 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 1.3 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 3.0 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 1.4 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 4.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 2.8 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 1.4 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 1.5 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 0.6 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 2.2 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 0.9 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.1 | 1.0 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.1 | 1.0 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 0.9 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 2.7 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.4 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 2.8 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.1 | 0.8 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 1.6 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 4.1 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.1 | 5.8 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 0.4 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.5 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 2.2 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 1.6 | GO:0035380 | C-3 sterol dehydrogenase (C-4 sterol decarboxylase) activity(GO:0000252) mevaldate reductase activity(GO:0004495) gluconate dehydrogenase activity(GO:0008875) epoxide dehydrogenase activity(GO:0018451) 5-exo-hydroxycamphor dehydrogenase activity(GO:0018452) 2-hydroxytetrahydrofuran dehydrogenase activity(GO:0018453) acetoin dehydrogenase activity(GO:0019152) phenylcoumaran benzylic ether reductase activity(GO:0032442) D-xylose:NADP reductase activity(GO:0032866) L-arabinose:NADP reductase activity(GO:0032867) D-arabinitol dehydrogenase, D-ribulose forming (NADP+) activity(GO:0033709) (R)-(-)-1,2,3,4-tetrahydronaphthol dehydrogenase activity(GO:0034831) 3-hydroxymenthone dehydrogenase activity(GO:0034840) very long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0035380) dihydrotestosterone 17-beta-dehydrogenase activity(GO:0035410) (R)-2-hydroxyisocaproate dehydrogenase activity(GO:0043713) L-arabinose 1-dehydrogenase (NADP+) activity(GO:0044103) L-xylulose reductase (NAD+) activity(GO:0044105) 3-ketoglucose-reductase activity(GO:0048258) D-arabinitol dehydrogenase, D-xylulose forming (NADP+) activity(GO:0052677) |
| 0.1 | 0.2 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.1 | 1.0 | GO:0001848 | complement binding(GO:0001848) |
| 0.1 | 0.5 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 6.3 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.1 | 1.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.9 | GO:0034951 | pivalyl-CoA mutase activity(GO:0034784) o-hydroxylaminobenzoate mutase activity(GO:0034951) lupeol synthase activity(GO:0042299) beta-amyrin synthase activity(GO:0042300) baruol synthase activity(GO:0080011) |
| 0.1 | 0.2 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.1 | 1.3 | GO:0008143 | poly(A) binding(GO:0008143) poly(U) RNA binding(GO:0008266) |
| 0.1 | 13.7 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 13.7 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.1 | 1.2 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 4.1 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 0.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.6 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 1.0 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.1 | 0.8 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.1 | 1.0 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 1.3 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 0.6 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 1.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.7 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 2.7 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 3.0 | GO:0008186 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 1.2 | GO:0008200 | ion channel inhibitor activity(GO:0008200) |
| 0.0 | 0.4 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.6 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 1.2 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.3 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.6 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 1.0 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 6.0 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.5 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 2.6 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.2 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 2.3 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 8.7 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.7 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.4 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.6 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 1.6 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 0.3 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.4 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.4 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.2 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.5 | GO:0061650 | ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.0 | 0.4 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.0 | 0.7 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 1.0 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 1.2 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.6 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 1.0 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 5.4 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 2.3 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.8 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 1.0 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.2 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.5 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.8 | GO:0004120 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) photoreceptor cyclic-nucleotide phosphodiesterase activity(GO:0004120) 7,8-dihydro-D-neopterin 2',3'-cyclic phosphate phosphodiesterase activity(GO:0044688) inositol phosphosphingolipid phospholipase activity(GO:0052712) inositol phosphorylceramide phospholipase activity(GO:0052713) mannosyl-inositol phosphorylceramide phospholipase activity(GO:0052714) mannosyl-diinositol phosphorylceramide phospholipase activity(GO:0052715) |
| 0.0 | 1.4 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 4.1 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.5 | GO:1990782 | protein tyrosine kinase binding(GO:1990782) |
| 0.0 | 0.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.2 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.2 | GO:0008932 | lytic endotransglycosylase activity(GO:0008932) lytic transglycosylase activity(GO:0008933) |
| 0.0 | 0.1 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.5 | GO:0003743 | translation initiation factor activity(GO:0003743) |