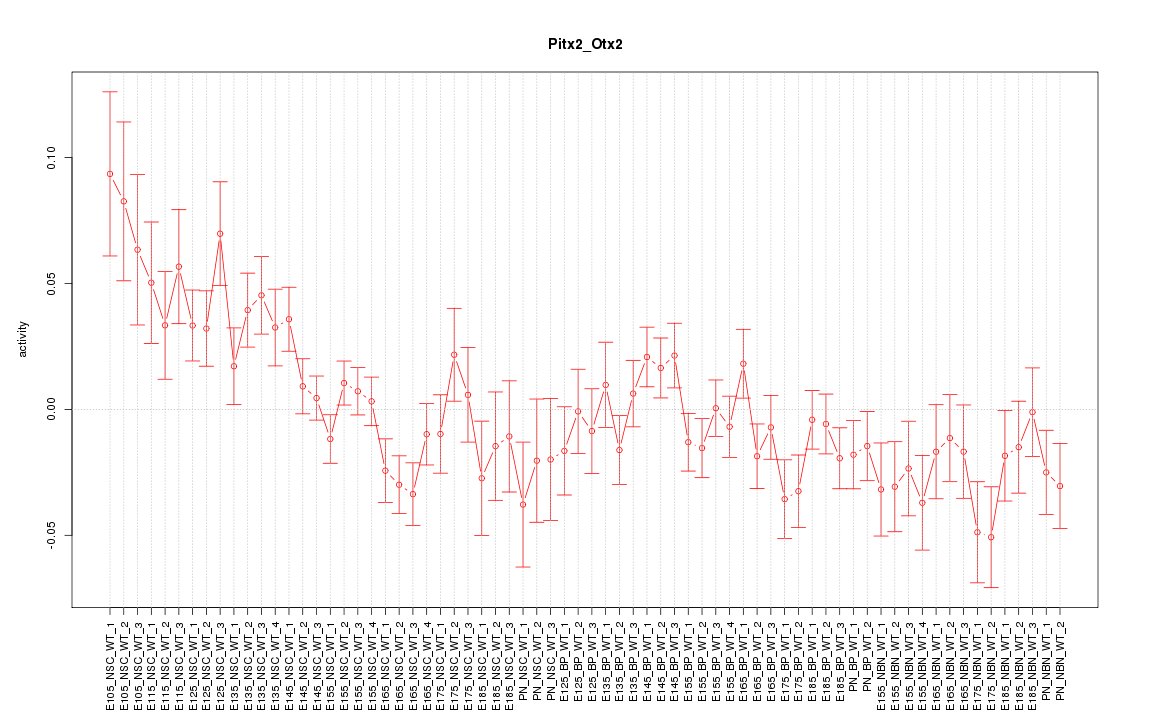
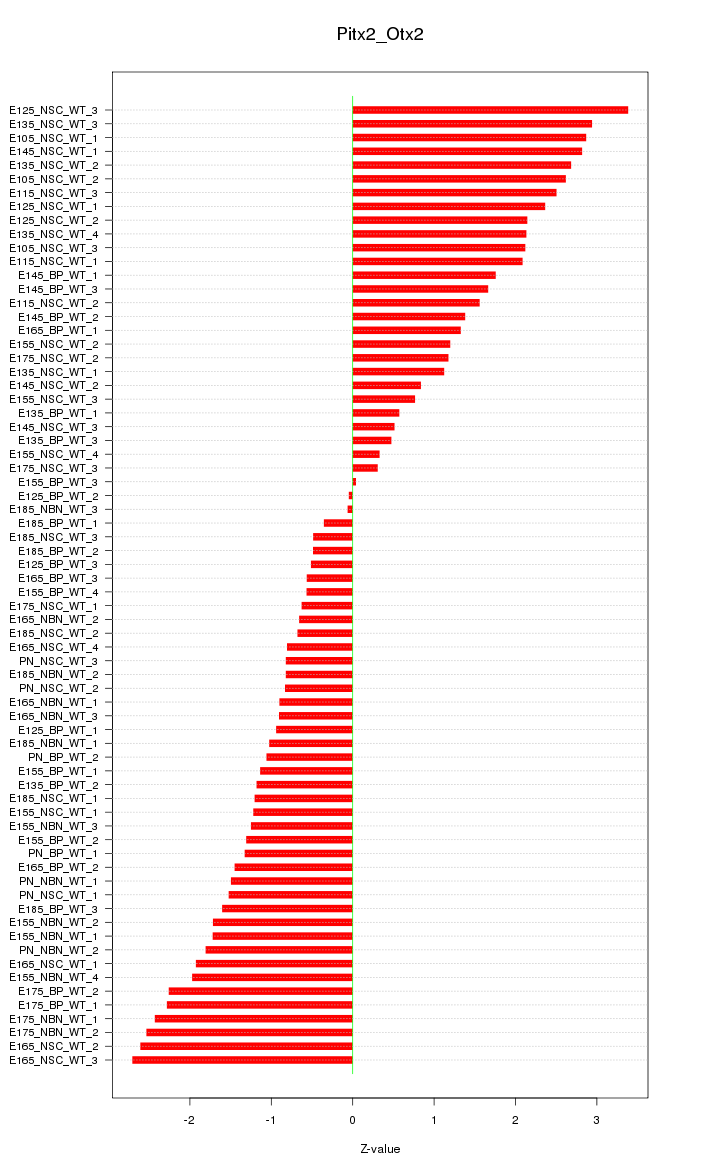
Motif ID: Pitx2_Otx2
Z-value: 1.614

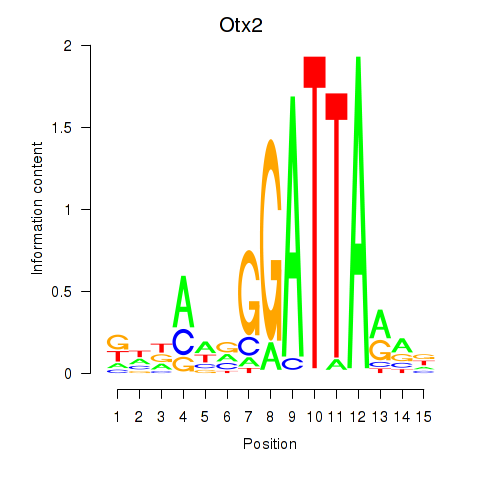
Transcription factors associated with Pitx2_Otx2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Otx2 | ENSMUSG00000021848.9 | Otx2 |
| Pitx2 | ENSMUSG00000028023.10 | Pitx2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Otx2 | mm10_v2_chr14_-_48662740_48662872 | 0.70 | 9.9e-12 | Click! |
| Pitx2 | mm10_v2_chr3_+_129199878_129199913 | 0.66 | 7.3e-10 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.7 | 17.1 | GO:0015938 | coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) |
| 5.6 | 16.8 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 5.0 | 14.9 | GO:0016095 | polyprenol catabolic process(GO:0016095) terpenoid catabolic process(GO:0016115) primary alcohol catabolic process(GO:0034310) |
| 4.0 | 12.0 | GO:0009177 | deoxyribonucleoside monophosphate biosynthetic process(GO:0009157) pyrimidine deoxyribonucleoside monophosphate biosynthetic process(GO:0009177) |
| 3.6 | 10.8 | GO:0021557 | oculomotor nerve development(GO:0021557) ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 3.2 | 12.7 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 2.9 | 8.7 | GO:0046166 | glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 2.9 | 14.3 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 2.8 | 13.8 | GO:0015705 | iodide transport(GO:0015705) |
| 2.6 | 7.7 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 2.4 | 7.3 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 2.4 | 7.1 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 2.3 | 4.6 | GO:0090202 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 2.1 | 8.6 | GO:1902990 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 2.1 | 6.4 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 2.1 | 6.3 | GO:0002765 | immune response-inhibiting signal transduction(GO:0002765) |
| 2.0 | 6.1 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) odontoblast differentiation(GO:0071895) regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 2.0 | 3.9 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 1.9 | 1.9 | GO:0071726 | response to diacyl bacterial lipopeptide(GO:0071724) cellular response to diacyl bacterial lipopeptide(GO:0071726) |
| 1.8 | 5.4 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 1.8 | 5.3 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 1.7 | 3.5 | GO:1903519 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 1.7 | 5.1 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 1.7 | 22.0 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 1.7 | 5.0 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 1.7 | 8.3 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 1.6 | 4.9 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 1.6 | 4.8 | GO:0050787 | glycolate metabolic process(GO:0009441) enzyme active site formation via L-cysteine sulfinic acid(GO:0018323) primary alcohol biosynthetic process(GO:0034309) cellular response to glyoxal(GO:0036471) glycolate biosynthetic process(GO:0046295) detoxification of mercury ion(GO:0050787) negative regulation of TRAIL-activated apoptotic signaling pathway(GO:1903122) regulation of pyrroline-5-carboxylate reductase activity(GO:1903167) positive regulation of pyrroline-5-carboxylate reductase activity(GO:1903168) regulation of tyrosine 3-monooxygenase activity(GO:1903176) positive regulation of tyrosine 3-monooxygenase activity(GO:1903178) L-dopa metabolic process(GO:1903184) L-dopa biosynthetic process(GO:1903185) glyoxal metabolic process(GO:1903189) glyoxal catabolic process(GO:1903190) regulation of L-dopa biosynthetic process(GO:1903195) positive regulation of L-dopa biosynthetic process(GO:1903197) regulation of L-dopa decarboxylase activity(GO:1903198) positive regulation of L-dopa decarboxylase activity(GO:1903200) positive regulation of cellular amino acid biosynthetic process(GO:2000284) positive regulation of androgen receptor activity(GO:2000825) |
| 1.6 | 9.4 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 1.6 | 4.7 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 1.6 | 4.7 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 1.5 | 4.5 | GO:0015920 | regulation of phosphatidylcholine catabolic process(GO:0010899) lipopolysaccharide transport(GO:0015920) |
| 1.4 | 4.3 | GO:0036292 | DNA rewinding(GO:0036292) |
| 1.4 | 7.2 | GO:0019249 | lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) |
| 1.4 | 11.6 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 1.4 | 5.8 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 1.4 | 8.6 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 1.4 | 7.1 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 1.3 | 4.0 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 1.3 | 6.7 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 1.3 | 6.6 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 1.3 | 32.0 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 1.3 | 5.1 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 1.3 | 5.1 | GO:0002339 | B cell selection(GO:0002339) |
| 1.2 | 12.5 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 1.2 | 3.7 | GO:0007525 | somatic muscle development(GO:0007525) |
| 1.2 | 7.4 | GO:0046502 | uroporphyrinogen III metabolic process(GO:0046502) |
| 1.2 | 3.6 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 1.2 | 12.0 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 1.2 | 3.5 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 1.2 | 6.9 | GO:0003383 | apical constriction(GO:0003383) |
| 1.1 | 7.8 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 1.1 | 5.5 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 1.0 | 6.0 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 1.0 | 4.9 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.9 | 3.6 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.9 | 2.7 | GO:0031627 | telomeric loop formation(GO:0031627) |
| 0.9 | 3.6 | GO:0046210 | nitrate catabolic process(GO:0043602) nitric oxide catabolic process(GO:0046210) cellular organohalogen metabolic process(GO:0090345) cellular organofluorine metabolic process(GO:0090346) |
| 0.9 | 2.7 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.9 | 3.5 | GO:0006203 | dGTP catabolic process(GO:0006203) dATP catabolic process(GO:0046061) |
| 0.9 | 2.6 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.8 | 4.1 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.8 | 6.5 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.8 | 2.4 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.8 | 6.3 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.8 | 3.1 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.7 | 3.0 | GO:0019614 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.7 | 2.9 | GO:0042462 | photoreceptor cell development(GO:0042461) eye photoreceptor cell development(GO:0042462) |
| 0.7 | 8.1 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 0.7 | 2.9 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.7 | 2.1 | GO:0015960 | diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) |
| 0.7 | 7.0 | GO:0018065 | protein-cofactor linkage(GO:0018065) |
| 0.7 | 10.4 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.7 | 2.8 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.7 | 5.5 | GO:0033504 | floor plate development(GO:0033504) |
| 0.7 | 3.4 | GO:0045918 | negative regulation of cytolysis(GO:0045918) |
| 0.7 | 8.2 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.7 | 8.2 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.7 | 19.2 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.7 | 1.3 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.7 | 2.0 | GO:0045472 | response to ether(GO:0045472) |
| 0.7 | 2.0 | GO:0006407 | rRNA export from nucleus(GO:0006407) |
| 0.6 | 3.2 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 0.6 | 3.2 | GO:0090666 | telomere assembly(GO:0032202) scaRNA localization to Cajal body(GO:0090666) |
| 0.6 | 1.9 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.6 | 5.6 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.6 | 2.5 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.6 | 3.1 | GO:0033087 | negative regulation of immature T cell proliferation(GO:0033087) |
| 0.6 | 3.7 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.6 | 1.8 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.6 | 1.8 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.6 | 3.5 | GO:0032460 | negative regulation of protein oligomerization(GO:0032460) negative regulation of protein homooligomerization(GO:0032463) |
| 0.6 | 2.3 | GO:0014012 | negative regulation of Schwann cell proliferation(GO:0010626) peripheral nervous system axon regeneration(GO:0014012) |
| 0.6 | 5.7 | GO:0006271 | DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.6 | 2.8 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.6 | 3.9 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.6 | 5.6 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.6 | 1.7 | GO:0009071 | serine family amino acid catabolic process(GO:0009071) |
| 0.5 | 2.2 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.5 | 6.0 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.5 | 1.6 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.5 | 3.2 | GO:0070814 | hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.5 | 1.6 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.5 | 2.6 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.5 | 2.6 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.5 | 1.5 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.5 | 1.5 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.5 | 1.5 | GO:0036118 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.5 | 1.5 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.5 | 7.0 | GO:0046697 | decidualization(GO:0046697) |
| 0.5 | 3.0 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.5 | 3.0 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.5 | 4.0 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.5 | 6.0 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.5 | 8.4 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.5 | 2.9 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.5 | 1.5 | GO:0050929 | corticospinal neuron axon guidance through spinal cord(GO:0021972) positive regulation of negative chemotaxis(GO:0050924) induction of negative chemotaxis(GO:0050929) negative regulation of mononuclear cell migration(GO:0071676) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.5 | 4.3 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.5 | 1.9 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.5 | 8.0 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.5 | 1.4 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.5 | 5.2 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.5 | 2.8 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.5 | 2.3 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.5 | 3.2 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.4 | 4.5 | GO:0046036 | GTP biosynthetic process(GO:0006183) CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.4 | 1.3 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.4 | 2.7 | GO:1902410 | mitotic cytokinetic process(GO:1902410) |
| 0.4 | 1.3 | GO:0000059 | protein import into nucleus, docking(GO:0000059) |
| 0.4 | 3.9 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.4 | 1.7 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.4 | 3.0 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.4 | 2.9 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) |
| 0.4 | 2.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.4 | 3.8 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.4 | 1.7 | GO:0031424 | keratinization(GO:0031424) |
| 0.4 | 2.5 | GO:0001980 | regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.4 | 1.2 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.4 | 1.2 | GO:0052428 | modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.4 | 2.0 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.4 | 1.6 | GO:0035356 | positive regulation of sequestering of triglyceride(GO:0010890) cellular triglyceride homeostasis(GO:0035356) |
| 0.4 | 9.2 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.4 | 2.0 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.4 | 0.8 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.4 | 2.4 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.4 | 3.1 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.4 | 3.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.4 | 5.0 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.4 | 6.5 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.4 | 2.7 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.4 | 3.0 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.4 | 5.7 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.4 | 2.2 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.4 | 5.6 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.4 | 1.5 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.4 | 13.6 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.4 | 4.4 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.4 | 3.3 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.4 | 1.4 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.4 | 1.1 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.4 | 2.5 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.4 | 1.1 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.4 | 3.6 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.4 | 1.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.4 | 1.1 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.4 | 2.1 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.3 | 1.0 | GO:2000152 | regulation of ubiquitin-specific protease activity(GO:2000152) |
| 0.3 | 2.4 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.3 | 1.4 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.3 | 1.4 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.3 | 0.3 | GO:0006702 | androgen biosynthetic process(GO:0006702) |
| 0.3 | 1.7 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.3 | 2.3 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.3 | 3.0 | GO:0000963 | mitochondrial RNA processing(GO:0000963) |
| 0.3 | 1.0 | GO:0051155 | positive regulation of myotube differentiation(GO:0010831) positive regulation of striated muscle cell differentiation(GO:0051155) |
| 0.3 | 1.0 | GO:0032513 | negative regulation of protein phosphatase type 2B activity(GO:0032513) |
| 0.3 | 3.9 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 0.3 | 2.3 | GO:0051974 | negative regulation of telomerase activity(GO:0051974) |
| 0.3 | 7.1 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.3 | 1.3 | GO:0033580 | protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.3 | 3.2 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.3 | 3.1 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.3 | 1.2 | GO:0032229 | negative regulation of synaptic transmission, GABAergic(GO:0032229) |
| 0.3 | 3.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.3 | 4.7 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.3 | 5.0 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.3 | 0.9 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.3 | 3.7 | GO:0036035 | osteoclast development(GO:0036035) |
| 0.3 | 3.7 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.3 | 1.5 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.3 | 5.4 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.3 | 1.2 | GO:0010726 | positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.3 | 7.4 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.3 | 3.5 | GO:0015816 | glycine transport(GO:0015816) |
| 0.3 | 0.9 | GO:0036166 | phenotypic switching(GO:0036166) |
| 0.3 | 1.2 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.3 | 1.1 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.3 | 2.0 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.3 | 2.3 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.3 | 4.1 | GO:0006085 | acetyl-CoA biosynthetic process(GO:0006085) |
| 0.3 | 6.0 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.3 | 6.2 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.3 | 0.8 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.3 | 1.9 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.3 | 5.9 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.3 | 3.7 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.3 | 0.3 | GO:0045410 | positive regulation of interleukin-6 biosynthetic process(GO:0045410) |
| 0.3 | 6.3 | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.3 | 1.1 | GO:0036506 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.3 | 5.0 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.3 | 13.9 | GO:0048538 | thymus development(GO:0048538) |
| 0.3 | 1.6 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.3 | 2.3 | GO:0032988 | ribonucleoprotein complex disassembly(GO:0032988) |
| 0.3 | 3.1 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.3 | 4.1 | GO:0060065 | uterus development(GO:0060065) |
| 0.3 | 1.5 | GO:0015817 | histidine transport(GO:0015817) |
| 0.3 | 3.6 | GO:2000010 | positive regulation of protein localization to cell surface(GO:2000010) |
| 0.3 | 1.5 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.3 | 6.1 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.3 | 4.1 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.3 | 1.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.2 | 2.7 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.2 | 1.5 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.2 | 4.1 | GO:1903427 | negative regulation of reactive oxygen species biosynthetic process(GO:1903427) |
| 0.2 | 1.4 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.2 | 9.1 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.2 | 4.5 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.2 | 5.4 | GO:0042168 | heme metabolic process(GO:0042168) |
| 0.2 | 3.3 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.2 | 1.4 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.2 | 2.8 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.2 | 2.8 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.2 | 2.1 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.2 | 2.1 | GO:0055070 | copper ion homeostasis(GO:0055070) |
| 0.2 | 1.8 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.2 | 2.0 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.2 | 7.0 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.2 | 1.5 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.2 | 0.2 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.2 | 2.3 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.2 | 0.6 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.2 | 1.6 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.2 | 5.9 | GO:0007094 | mitotic spindle assembly checkpoint(GO:0007094) |
| 0.2 | 1.2 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.2 | 0.6 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.2 | 1.0 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 0.2 | 10.8 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.2 | 1.9 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.2 | 3.0 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.2 | 2.6 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.2 | 1.1 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.2 | 3.0 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.2 | 4.0 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.2 | 22.8 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.2 | 0.9 | GO:0048318 | axial mesoderm development(GO:0048318) |
| 0.2 | 1.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.2 | 0.9 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.2 | 2.1 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.2 | 1.9 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.2 | 1.9 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.2 | 0.7 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) cellular response to X-ray(GO:0071481) |
| 0.2 | 2.5 | GO:0060135 | maternal process involved in female pregnancy(GO:0060135) |
| 0.2 | 2.3 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.2 | 1.0 | GO:1903867 | chorion development(GO:0060717) extraembryonic membrane development(GO:1903867) |
| 0.2 | 0.5 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.2 | 2.1 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.2 | 4.4 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.2 | 0.5 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.2 | 4.7 | GO:0016445 | somatic diversification of immunoglobulins(GO:0016445) |
| 0.2 | 1.4 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.2 | 1.1 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.2 | 4.0 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.2 | 1.7 | GO:0035413 | positive regulation of catenin import into nucleus(GO:0035413) |
| 0.2 | 0.6 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.2 | 5.8 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.2 | 4.9 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.2 | 2.1 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.1 | 2.2 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.1 | 7.5 | GO:0048146 | positive regulation of fibroblast proliferation(GO:0048146) |
| 0.1 | 1.6 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 2.1 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 2.2 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.1 | 0.7 | GO:0009650 | UV protection(GO:0009650) |
| 0.1 | 1.0 | GO:1901799 | negative regulation of proteasomal protein catabolic process(GO:1901799) |
| 0.1 | 1.4 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.1 | 1.1 | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) |
| 0.1 | 0.8 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.4 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.1 | 1.6 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 0.9 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.1 | 0.3 | GO:0036337 | Fas signaling pathway(GO:0036337) |
| 0.1 | 1.2 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 0.3 | GO:0032466 | negative regulation of cytokinesis(GO:0032466) |
| 0.1 | 1.3 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 14.1 | GO:0031109 | microtubule polymerization or depolymerization(GO:0031109) |
| 0.1 | 1.9 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 1.7 | GO:0010388 | protein deneddylation(GO:0000338) cullin deneddylation(GO:0010388) |
| 0.1 | 0.4 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.1 | 1.2 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.1 | 1.8 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.1 | 1.1 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.1 | 8.5 | GO:0007088 | regulation of mitotic nuclear division(GO:0007088) |
| 0.1 | 6.6 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.1 | 1.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.1 | 0.7 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.1 | 1.5 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.1 | 2.2 | GO:0006582 | melanin metabolic process(GO:0006582) melanin biosynthetic process(GO:0042438) |
| 0.1 | 0.6 | GO:0018002 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.1 | 1.1 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.1 | 0.3 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.1 | 1.1 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.1 | 1.6 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 0.7 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.1 | 0.5 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.1 | 6.0 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.1 | 2.2 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 0.7 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.1 | 0.8 | GO:0001996 | regulation of systemic arterial blood pressure by norepinephrine-epinephrine(GO:0001993) positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) |
| 0.1 | 0.3 | GO:0090343 | positive regulation of cell aging(GO:0090343) positive regulation of cellular senescence(GO:2000774) |
| 0.1 | 2.2 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 1.4 | GO:0042274 | ribosomal small subunit biogenesis(GO:0042274) |
| 0.1 | 5.9 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.1 | 3.8 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) |
| 0.1 | 2.5 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.1 | 1.8 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.1 | 0.7 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.1 | 0.5 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.1 | 1.1 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.1 | 2.2 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 1.7 | GO:0030201 | heparan sulfate proteoglycan metabolic process(GO:0030201) |
| 0.1 | 1.2 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.1 | 0.6 | GO:0032691 | negative regulation of interleukin-1 beta production(GO:0032691) |
| 0.1 | 2.5 | GO:0043039 | tRNA aminoacylation for protein translation(GO:0006418) tRNA aminoacylation(GO:0043039) |
| 0.1 | 1.4 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.1 | 1.5 | GO:0030336 | negative regulation of cell migration(GO:0030336) negative regulation of cell motility(GO:2000146) |
| 0.1 | 0.9 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.1 | 1.9 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.1 | 4.2 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.1 | 0.7 | GO:0021554 | optic nerve development(GO:0021554) |
| 0.1 | 2.0 | GO:0034724 | DNA replication-independent nucleosome assembly(GO:0006336) DNA replication-independent nucleosome organization(GO:0034724) |
| 0.1 | 2.6 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 0.4 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.1 | 0.2 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 3.4 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 1.2 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 | 3.3 | GO:0000725 | double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 0.1 | 1.5 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.1 | 0.4 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 3.6 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.1 | 1.0 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.1 | 3.1 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.1 | 0.5 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.1 | 3.3 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.1 | 0.9 | GO:0006625 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.1 | 0.5 | GO:0002507 | tolerance induction(GO:0002507) |
| 0.1 | 1.6 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.1 | 3.1 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.1 | 0.4 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.1 | 1.9 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.1 | 0.5 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.1 | 0.2 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.1 | 0.5 | GO:0098656 | anion transmembrane transport(GO:0098656) |
| 0.1 | 6.3 | GO:0098780 | mitophagy in response to mitochondrial depolarization(GO:0098779) response to mitochondrial depolarisation(GO:0098780) |
| 0.1 | 0.3 | GO:0035428 | hexose transmembrane transport(GO:0035428) |
| 0.1 | 0.4 | GO:0045056 | transcytosis(GO:0045056) |
| 0.1 | 0.3 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.1 | 0.6 | GO:0001895 | retina homeostasis(GO:0001895) |
| 0.0 | 1.2 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 1.2 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.0 | 0.4 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.0 | 0.4 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.0 | 0.8 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.4 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.0 | 1.3 | GO:0006006 | glucose metabolic process(GO:0006006) |
| 0.0 | 1.1 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 0.3 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.4 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 1.8 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.0 | 0.9 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.4 | GO:0035646 | endosome to melanosome transport(GO:0035646) endosome to pigment granule transport(GO:0043485) pigment granule maturation(GO:0048757) |
| 0.0 | 0.1 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.0 | 2.3 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.2 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 5.0 | GO:0000398 | RNA splicing, via transesterification reactions(GO:0000375) RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 2.1 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.6 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.2 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.0 | 0.2 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.2 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.5 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 1.0 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.2 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.0 | 1.1 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.2 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.4 | GO:0045333 | cellular respiration(GO:0045333) |
| 0.0 | 0.4 | GO:0003229 | ventricular cardiac muscle tissue development(GO:0003229) |
| 0.0 | 1.3 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.0 | 0.1 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.0 | 0.1 | GO:0030836 | positive regulation of actin filament depolymerization(GO:0030836) positive regulation of protein depolymerization(GO:1901881) |
| 0.0 | 0.1 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.2 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.4 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.9 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.3 | GO:0048709 | oligodendrocyte differentiation(GO:0048709) |
| 0.0 | 0.1 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 26.7 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 2.8 | 8.3 | GO:1990393 | 3M complex(GO:1990393) |
| 2.7 | 8.2 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 2.6 | 7.8 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 2.3 | 16.3 | GO:0001940 | male pronucleus(GO:0001940) |
| 1.8 | 7.2 | GO:0032021 | NELF complex(GO:0032021) |
| 1.7 | 5.0 | GO:0038045 | large latent transforming growth factor-beta complex(GO:0038045) |
| 1.6 | 4.9 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 1.6 | 6.5 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 1.6 | 7.8 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 1.5 | 24.8 | GO:0005652 | nuclear lamina(GO:0005652) |
| 1.4 | 11.6 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 1.4 | 4.3 | GO:1990047 | spindle matrix(GO:1990047) |
| 1.3 | 4.0 | GO:0071914 | prominosome(GO:0071914) |
| 1.3 | 6.7 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 1.3 | 21.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 1.1 | 2.3 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 1.1 | 11.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 1.1 | 3.2 | GO:0000811 | GINS complex(GO:0000811) |
| 1.0 | 3.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.9 | 4.7 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.9 | 3.6 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.9 | 5.4 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.9 | 5.4 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.9 | 2.7 | GO:0030690 | Noc1p-Noc2p complex(GO:0030690) |
| 0.9 | 7.9 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.9 | 6.9 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.8 | 8.0 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.8 | 10.1 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.7 | 9.5 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.7 | 3.6 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.7 | 1.4 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.7 | 6.3 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.7 | 8.4 | GO:0031105 | septin complex(GO:0031105) |
| 0.7 | 8.9 | GO:0032797 | SMN complex(GO:0032797) |
| 0.7 | 5.5 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.7 | 2.0 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.7 | 4.6 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.7 | 32.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.6 | 3.9 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.6 | 1.9 | GO:1990879 | CST complex(GO:1990879) |
| 0.6 | 3.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.6 | 5.6 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.6 | 43.8 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.6 | 3.6 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.6 | 8.3 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.6 | 8.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.6 | 6.4 | GO:0070938 | contractile ring(GO:0070938) |
| 0.6 | 2.3 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.6 | 1.7 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.6 | 6.1 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.5 | 7.1 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.5 | 2.7 | GO:0008623 | CHRAC(GO:0008623) |
| 0.5 | 2.1 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.5 | 2.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.5 | 2.0 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.5 | 40.8 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.5 | 7.3 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.5 | 6.7 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.5 | 1.9 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.5 | 3.3 | GO:0030677 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.5 | 12.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.5 | 1.8 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.5 | 3.2 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.5 | 6.3 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.4 | 2.5 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.4 | 1.7 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.4 | 4.6 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.4 | 5.8 | GO:0043218 | compact myelin(GO:0043218) |
| 0.4 | 0.7 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.4 | 1.8 | GO:0090543 | Flemming body(GO:0090543) |
| 0.4 | 0.7 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.3 | 1.0 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.3 | 1.0 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.3 | 3.0 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.3 | 3.0 | GO:0000243 | commitment complex(GO:0000243) |
| 0.3 | 13.8 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.3 | 1.9 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.3 | 5.3 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.3 | 2.5 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.3 | 4.5 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.3 | 2.1 | GO:0070187 | telosome(GO:0070187) |
| 0.3 | 0.9 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.3 | 2.0 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.3 | 5.2 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.3 | 16.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.3 | 4.0 | GO:0032039 | integrator complex(GO:0032039) |
| 0.3 | 2.8 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.3 | 2.0 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.3 | 4.5 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.3 | 3.1 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.3 | 0.5 | GO:0098576 | lumenal side of membrane(GO:0098576) |
| 0.3 | 1.1 | GO:0030894 | replisome(GO:0030894) |
| 0.3 | 2.1 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.3 | 12.3 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.3 | 8.0 | GO:0015030 | Cajal body(GO:0015030) |
| 0.2 | 1.0 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.2 | 1.2 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.2 | 9.2 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.2 | 1.4 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.2 | 1.6 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.2 | 2.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.2 | 2.0 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.2 | 8.8 | GO:1990204 | oxidoreductase complex(GO:1990204) |
| 0.2 | 1.5 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.2 | 3.4 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.2 | 1.2 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.2 | 15.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.2 | 9.7 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.2 | 1.2 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.2 | 3.4 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.2 | 0.6 | GO:0031415 | NatA complex(GO:0031415) |
| 0.2 | 0.6 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.2 | 1.9 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.2 | 13.1 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.2 | 0.7 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.2 | 1.7 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.2 | 2.1 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.2 | 8.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.2 | 1.5 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.2 | 5.8 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.2 | 1.3 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.2 | 5.0 | GO:0097223 | sperm part(GO:0097223) |
| 0.2 | 3.3 | GO:0030684 | preribosome(GO:0030684) |
| 0.2 | 1.1 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.2 | 1.5 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.2 | 0.5 | GO:0000801 | central element(GO:0000801) |
| 0.2 | 5.2 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.2 | 1.1 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.1 | 12.6 | GO:0005840 | ribosome(GO:0005840) |
| 0.1 | 8.6 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 2.8 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 8.0 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 4.9 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 1.0 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.1 | 1.6 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 1.8 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 2.5 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 1.6 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 14.1 | GO:0005903 | brush border(GO:0005903) |
| 0.1 | 9.1 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 0.9 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.1 | 2.4 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 0.6 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.1 | 2.0 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 1.5 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 1.9 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 1.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 1.0 | GO:0046930 | pore complex(GO:0046930) Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 6.4 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 7.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 0.7 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.1 | 1.2 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 1.6 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.1 | 1.3 | GO:0033202 | DNA helicase complex(GO:0033202) |
| 0.1 | 0.4 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.1 | 7.8 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 1.1 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 2.2 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.1 | 2.3 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 2.2 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 0.3 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.1 | 0.6 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 1.6 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.1 | 15.8 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 0.8 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 2.2 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 0.6 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 0.5 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 0.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 0.6 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 1.3 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 2.2 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 3.6 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 7.8 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 1.4 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 7.2 | GO:0000776 | kinetochore(GO:0000776) |
| 0.1 | 0.4 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 5.1 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.1 | 0.6 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 0.5 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 0.9 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 2.2 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 1.0 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 0.3 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 2.0 | GO:0032592 | integral component of mitochondrial membrane(GO:0032592) |
| 0.1 | 2.4 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 3.8 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 4.4 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 5.5 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 8.4 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 0.5 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 1.6 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 12.2 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 2.4 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 2.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.6 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 1.2 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 1.2 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.0 | 1.3 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.3 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 1.1 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 1.1 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 1.3 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 11.9 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 2.1 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.5 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 1.6 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 65.0 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 0.9 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 1.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 1.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.7 | 17.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 4.6 | 13.7 | GO:0070905 | serine binding(GO:0070905) |
| 3.5 | 13.8 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 2.8 | 22.0 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 2.7 | 8.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 2.5 | 14.9 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 2.4 | 7.1 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 2.1 | 2.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 1.9 | 7.8 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 1.9 | 5.8 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 1.9 | 7.5 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 1.8 | 7.2 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 1.8 | 8.9 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 1.7 | 5.1 | GO:0018738 | S-formylglutathione hydrolase activity(GO:0018738) |
| 1.6 | 4.8 | GO:0044388 | tyrosine 3-monooxygenase activator activity(GO:0036470) L-dopa decarboxylase activator activity(GO:0036478) small protein activating enzyme binding(GO:0044388) cupric ion binding(GO:1903135) cuprous ion binding(GO:1903136) glyoxalase (glycolic acid-forming) activity(GO:1990422) |
| 1.6 | 12.7 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 1.6 | 6.3 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 1.6 | 4.7 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 1.5 | 4.6 | GO:0034618 | arginine binding(GO:0034618) |
| 1.5 | 4.5 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 1.5 | 6.0 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 1.4 | 4.3 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 1.4 | 6.9 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 1.3 | 3.9 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 1.3 | 6.4 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 1.2 | 5.0 | GO:0050436 | microfibril binding(GO:0050436) |
| 1.2 | 4.9 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 1.2 | 8.2 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 1.2 | 15.3 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 1.1 | 4.5 | GO:0035877 | death effector domain binding(GO:0035877) |
| 1.1 | 12.0 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 1.1 | 31.4 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 1.1 | 9.5 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 1.1 | 11.6 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 1.1 | 11.6 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 1.0 | 5.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 1.0 | 3.0 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 1.0 | 2.9 | GO:0070140 | isopeptidase activity(GO:0070122) ubiquitin-like protein-specific isopeptidase activity(GO:0070138) SUMO-specific isopeptidase activity(GO:0070140) |
| 0.9 | 3.8 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.9 | 6.5 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.9 | 3.6 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.9 | 12.6 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.9 | 4.5 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.9 | 3.6 | GO:0016708 | nitric oxide dioxygenase activity(GO:0008941) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of two atoms of oxygen into one donor(GO:0016708) iron-cytochrome-c reductase activity(GO:0047726) |
| 0.9 | 13.0 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.9 | 6.0 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.8 | 4.1 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.8 | 2.5 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.8 | 2.4 | GO:0004560 | alpha-L-fucosidase activity(GO:0004560) |
| 0.8 | 2.4 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
| 0.8 | 2.4 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.8 | 11.0 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.8 | 3.1 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.7 | 6.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.7 | 10.9 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.7 | 32.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.7 | 2.8 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.7 | 3.5 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.7 | 5.4 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.7 | 2.7 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.7 | 1.3 | GO:0008732 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.7 | 4.0 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.6 | 7.7 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.6 | 5.0 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.6 | 6.7 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.6 | 1.8 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.6 | 8.2 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.6 | 4.7 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.6 | 2.3 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.6 | 1.7 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.6 | 7.9 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.6 | 5.6 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.5 | 3.3 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.5 | 2.1 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.5 | 3.6 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.5 | 7.1 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.5 | 1.5 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.5 | 2.5 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.5 | 8.4 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.5 | 2.0 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.5 | 1.9 | GO:0004921 | interleukin-11 receptor activity(GO:0004921) interleukin-11 binding(GO:0019970) |
| 0.5 | 10.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.5 | 6.6 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.5 | 1.9 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.5 | 4.7 | GO:0034951 | pivalyl-CoA mutase activity(GO:0034784) o-hydroxylaminobenzoate mutase activity(GO:0034951) lupeol synthase activity(GO:0042299) beta-amyrin synthase activity(GO:0042300) baruol synthase activity(GO:0080011) |
| 0.5 | 1.4 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.5 | 4.1 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.5 | 10.8 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.5 | 1.8 | GO:0050347 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.4 | 1.3 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.4 | 2.2 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.4 | 3.5 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.4 | 4.0 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.4 | 0.4 | GO:0001083 | transcription factor activity, RNA polymerase II basal transcription factor binding(GO:0001083) |
| 0.4 | 2.5 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) |
| 0.4 | 2.8 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.4 | 1.2 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.4 | 1.9 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.4 | 1.5 | GO:0086038 | calcium:sodium antiporter activity involved in regulation of cardiac muscle cell membrane potential(GO:0086038) |
| 0.4 | 4.0 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.4 | 3.3 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.4 | 4.3 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.4 | 6.9 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.4 | 5.8 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.4 | 0.7 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.4 | 2.5 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.4 | 1.1 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.4 | 5.6 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.4 | 1.1 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.4 | 4.9 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.3 | 3.5 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.3 | 2.8 | GO:0008251 | tRNA-specific adenosine deaminase activity(GO:0008251) |
| 0.3 | 5.2 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.3 | 1.4 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.3 | 9.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.3 | 2.3 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.3 | 0.7 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.3 | 1.3 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.3 | 11.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.3 | 4.4 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.3 | 13.3 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.3 | 1.2 | GO:0004104 | cholinesterase activity(GO:0004104) choline binding(GO:0033265) |
| 0.3 | 5.5 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.3 | 1.5 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.3 | 65.9 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.3 | 4.5 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.3 | 0.9 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.3 | 6.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.3 | 1.2 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) |
| 0.3 | 7.4 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.3 | 3.3 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.3 | 2.0 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.3 | 1.4 | GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity(GO:0034602) |
| 0.3 | 1.4 | GO:0008808 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) |
| 0.3 | 2.8 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.3 | 0.8 | GO:0048039 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) ubiquinone binding(GO:0048039) |
| 0.3 | 0.8 | GO:0030362 | protein phosphatase type 4 regulator activity(GO:0030362) |
| 0.3 | 2.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.3 | 3.1 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.3 | 1.3 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.3 | 5.6 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.3 | 2.0 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.3 | 1.5 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) |
| 0.3 | 2.3 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.3 | 1.8 | GO:0015099 | nickel cation transmembrane transporter activity(GO:0015099) |
| 0.3 | 5.8 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 1.0 | GO:0016531 | metallochaperone activity(GO:0016530) copper chaperone activity(GO:0016531) |
| 0.2 | 7.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.2 | 2.9 | GO:0008430 | selenium binding(GO:0008430) |
| 0.2 | 2.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.2 | 0.9 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.2 | 1.9 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.2 | 0.9 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.2 | 2.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.2 | 5.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.2 | 2.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.2 | 3.4 | GO:0001848 | complement binding(GO:0001848) |
| 0.2 | 1.5 | GO:0045703 | pinocarveol dehydrogenase activity(GO:0018446) chloral hydrate dehydrogenase activity(GO:0018447) hydroxymethylmethylsilanediol oxidase activity(GO:0018448) 1-phenylethanol dehydrogenase activity(GO:0018449) myrtenol dehydrogenase activity(GO:0018450) cis-1,2-dihydroxy-1,2-dihydro-8-carboxynaphthalene dehydrogenase activity(GO:0034522) 3-hydroxy-4-methyloctanoyl-CoA dehydrogenase activity(GO:0034582) 2-hydroxy-4-isopropenylcyclohexane-1-carboxyl-CoA dehydrogenase activity(GO:0034778) cis-9,10-dihydroanthracene-9,10-diol dehydrogenase activity(GO:0034817) citronellol dehydrogenase activity(GO:0034821) naphthyl-2-hydroxymethyl-succinyl-CoA dehydrogenase activity(GO:0034847) 2,4,4-trimethyl-1-pentanol dehydrogenase activity(GO:0034863) 2,4,4-trimethyl-3-hydroxypentanoyl-CoA dehydrogenase activity(GO:0034868) 1-hydroxy-4,4-dimethylpentan-3-one dehydrogenase activity(GO:0034871) endosulfan diol dehydrogenase activity(GO:0034891) endosulfan hydroxyether dehydrogenase activity(GO:0034901) 3-hydroxy-2-methylhexanoyl-CoA dehydrogenase activity(GO:0034918) 3-hydroxy-2,6-dimethyl-5-methylene-heptanoyl-CoA dehydrogenase activity(GO:0034944) versicolorin reductase activity(GO:0042469) ketoreductase activity(GO:0045703) |
| 0.2 | 1.3 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.2 | 1.3 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.2 | 1.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.2 | 8.3 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.2 | 3.5 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.2 | 0.6 | GO:0008176 | tRNA (guanine-N7-)-methyltransferase activity(GO:0008176) |
| 0.2 | 1.4 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.2 | 2.0 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.2 | 2.8 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.2 | 3.7 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.2 | 1.9 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.2 | 3.6 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.2 | 1.7 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.2 | 2.4 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.2 | 7.0 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.2 | 4.4 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) |
| 0.2 | 3.2 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.2 | 3.0 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.2 | 2.1 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.2 | 7.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.2 | 1.0 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.2 | 3.0 | GO:0042165 | neurotransmitter binding(GO:0042165) |
| 0.2 | 3.8 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.2 | 1.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.2 | 0.5 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.2 | 0.8 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.2 | 1.0 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.2 | 4.1 | GO:0070717 | poly-purine tract binding(GO:0070717) |
| 0.2 | 0.5 | GO:0070890 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.1 | 1.6 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 3.8 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 1.0 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.1 | 2.3 | GO:0016594 | glycine binding(GO:0016594) |
| 0.1 | 0.4 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.1 | 0.6 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.1 | 1.7 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.4 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.1 | 5.1 | GO:0050694 | galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 2.7 | GO:0015923 | mannosidase activity(GO:0015923) |
| 0.1 | 4.8 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 3.3 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.1 | 0.6 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.1 | 4.6 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.1 | 0.3 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.1 | 0.9 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.1 | 3.8 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 1.0 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 1.8 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 6.0 | GO:0016651 | oxidoreductase activity, acting on NAD(P)H(GO:0016651) |
| 0.1 | 0.6 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.1 | 1.7 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.1 | 0.8 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.1 | 0.9 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.1 | 0.6 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.1 | 6.9 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 0.8 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.1 | 12.9 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.1 | 0.8 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 1.9 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.8 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 1.9 | GO:0016891 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) |
| 0.1 | 3.3 | GO:0018602 | sulfonate dioxygenase activity(GO:0000907) 2,4-dichlorophenoxyacetate alpha-ketoglutarate dioxygenase activity(GO:0018602) hypophosphite dioxygenase activity(GO:0034792) DNA-N1-methyladenine dioxygenase activity(GO:0043734) gibberellin 2-beta-dioxygenase activity(GO:0045543) C-19 gibberellin 2-beta-dioxygenase activity(GO:0052634) C-20 gibberellin 2-beta-dioxygenase activity(GO:0052635) |
| 0.1 | 3.6 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.1 | 0.9 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 1.0 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 4.0 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.1 | 0.8 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.1 | 1.4 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 1.5 | GO:0035380 | C-3 sterol dehydrogenase (C-4 sterol decarboxylase) activity(GO:0000252) mevaldate reductase activity(GO:0004495) gluconate dehydrogenase activity(GO:0008875) epoxide dehydrogenase activity(GO:0018451) 5-exo-hydroxycamphor dehydrogenase activity(GO:0018452) 2-hydroxytetrahydrofuran dehydrogenase activity(GO:0018453) acetoin dehydrogenase activity(GO:0019152) phenylcoumaran benzylic ether reductase activity(GO:0032442) D-xylose:NADP reductase activity(GO:0032866) L-arabinose:NADP reductase activity(GO:0032867) D-arabinitol dehydrogenase, D-ribulose forming (NADP+) activity(GO:0033709) (R)-(-)-1,2,3,4-tetrahydronaphthol dehydrogenase activity(GO:0034831) 3-hydroxymenthone dehydrogenase activity(GO:0034840) very long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0035380) dihydrotestosterone 17-beta-dehydrogenase activity(GO:0035410) (R)-2-hydroxyisocaproate dehydrogenase activity(GO:0043713) L-arabinose 1-dehydrogenase (NADP+) activity(GO:0044103) L-xylulose reductase (NAD+) activity(GO:0044105) 3-ketoglucose-reductase activity(GO:0048258) D-arabinitol dehydrogenase, D-xylulose forming (NADP+) activity(GO:0052677) |
| 0.1 | 0.3 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.1 | 0.7 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.1 | 1.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.2 | GO:0004936 | alpha-adrenergic receptor activity(GO:0004936) |
| 0.1 | 3.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 1.8 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 4.5 | GO:0008186 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.1 | 4.4 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 3.5 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 5.5 | GO:0004386 | helicase activity(GO:0004386) |
| 0.1 | 0.6 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 2.9 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.1 | 0.8 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 1.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.5 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.1 | 1.0 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 9.8 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 1.0 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.1 | 0.6 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 15.7 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.1 | 1.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) metalloexopeptidase activity(GO:0008235) |
| 0.1 | 1.0 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 0.4 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 1.7 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 52.0 | GO:0044822 | poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.6 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 17.0 | GO:0000987 | core promoter proximal region sequence-specific DNA binding(GO:0000987) |
| 0.0 | 2.0 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 3.6 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.2 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 1.3 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 4.0 | GO:0008237 | metallopeptidase activity(GO:0008237) |
| 0.0 | 0.7 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.5 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 1.1 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.5 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.7 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.8 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.4 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.6 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.3 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.3 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.3 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.8 | GO:0005402 | cation:sugar symporter activity(GO:0005402) |
| 0.0 | 1.1 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 2.0 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 1.2 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 3.6 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.3 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.4 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 1.4 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.3 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.3 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.7 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.1 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.0 | 0.5 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.1 | GO:0015295 | solute:proton symporter activity(GO:0015295) |
| 0.0 | 1.1 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.5 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 1.0 | GO:0015399 | primary active transmembrane transporter activity(GO:0015399) P-P-bond-hydrolysis-driven transmembrane transporter activity(GO:0015405) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 0.3 | GO:0001076 | transcription factor activity, RNA polymerase II transcription factor binding(GO:0001076) |
| 0.0 | 0.4 | GO:0052771 | coenzyme F390-A hydrolase activity(GO:0052770) coenzyme F390-G hydrolase activity(GO:0052771) |
| 0.0 | 1.0 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.7 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 2.1 | GO:0003682 | chromatin binding(GO:0003682) |