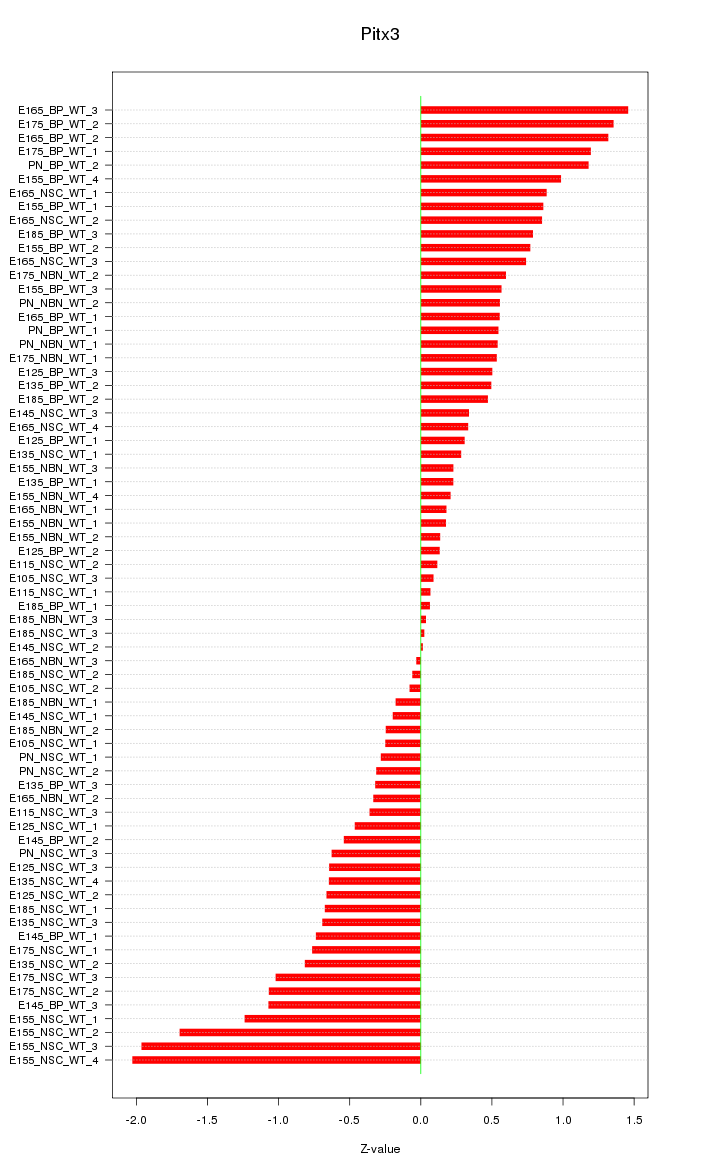
Motif ID: Pitx3
Z-value: 0.741
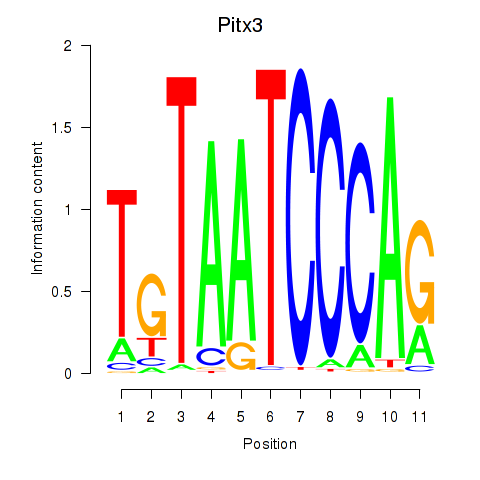
Transcription factors associated with Pitx3:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Pitx3 | ENSMUSG00000025229.9 | Pitx3 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.4 | GO:1990743 | protein sialylation(GO:1990743) |
| 0.8 | 4.9 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.8 | 3.9 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.7 | 2.1 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 0.6 | 1.9 | GO:0070944 | neutrophil mediated killing of bacterium(GO:0070944) |
| 0.6 | 1.2 | GO:2000110 | negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.6 | 2.3 | GO:0007066 | female meiosis sister chromatid cohesion(GO:0007066) |
| 0.5 | 0.5 | GO:0060854 | patterning of lymph vessels(GO:0060854) |
| 0.5 | 1.5 | GO:0042275 | error-free postreplication DNA repair(GO:0042275) |
| 0.5 | 2.5 | GO:0044838 | cell quiescence(GO:0044838) |
| 0.5 | 1.4 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.4 | 2.0 | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.4 | 1.9 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.4 | 1.5 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.3 | 1.3 | GO:0072362 | regulation of glycolytic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072362) |
| 0.3 | 0.9 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.3 | 1.2 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.3 | 1.7 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.3 | 1.3 | GO:0010359 | regulation of anion channel activity(GO:0010359) |
| 0.3 | 1.0 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.2 | 0.7 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.2 | 1.2 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.2 | 0.7 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.2 | 0.5 | GO:2000096 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.2 | 1.0 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.2 | 1.2 | GO:0051012 | microtubule sliding(GO:0051012) negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 0.2 | 0.9 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.2 | 1.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.2 | 1.0 | GO:0021764 | amygdala development(GO:0021764) |
| 0.2 | 0.6 | GO:1901731 | calcium-mediated signaling using extracellular calcium source(GO:0035585) positive regulation of platelet aggregation(GO:1901731) |
| 0.2 | 0.6 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.2 | 0.9 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.2 | 0.9 | GO:0006689 | ganglioside catabolic process(GO:0006689) oligosaccharide catabolic process(GO:0009313) |
| 0.2 | 0.9 | GO:2000587 | regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.2 | 0.5 | GO:0050929 | corticospinal neuron axon guidance through spinal cord(GO:0021972) positive regulation of negative chemotaxis(GO:0050924) induction of negative chemotaxis(GO:0050929) negative regulation of mononuclear cell migration(GO:0071676) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.2 | 1.0 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.2 | 0.8 | GO:2001168 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.2 | 0.5 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 0.2 | 0.6 | GO:1900272 | negative regulation of chronic inflammatory response(GO:0002677) negative regulation of long-term synaptic potentiation(GO:1900272) |
| 0.2 | 0.5 | GO:0000966 | RNA 5'-end processing(GO:0000966) |
| 0.1 | 0.1 | GO:0071033 | nuclear retention of pre-mRNA at the site of transcription(GO:0071033) |
| 0.1 | 0.4 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 | 0.4 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.1 | 0.7 | GO:0051189 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.1 | 0.6 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.1 | 0.4 | GO:0010871 | negative regulation of receptor biosynthetic process(GO:0010871) |
| 0.1 | 0.4 | GO:0016095 | polyprenol catabolic process(GO:0016095) terpenoid catabolic process(GO:0016115) primary alcohol catabolic process(GO:0034310) |
| 0.1 | 0.4 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.1 | 0.4 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.1 | 1.2 | GO:0060613 | fat pad development(GO:0060613) |
| 0.1 | 0.7 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.1 | 0.9 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.1 | 0.4 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.1 | 0.4 | GO:0070124 | mitochondrial translational initiation(GO:0070124) |
| 0.1 | 2.4 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.1 | 1.8 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.1 | 1.0 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 0.7 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 1.7 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.1 | 0.5 | GO:0036015 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.1 | 1.4 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.1 | 0.5 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.1 | 0.4 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 | 2.0 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.1 | 0.4 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.1 | 0.8 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.1 | 0.8 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 2.1 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.1 | 0.6 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 0.1 | 0.7 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 0.8 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.1 | 0.5 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.1 | 0.3 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.1 | 0.9 | GO:1901409 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 | 0.9 | GO:1900112 | regulation of histone H3-K9 trimethylation(GO:1900112) negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.1 | 0.7 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.1 | 0.7 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.1 | 1.9 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.1 | 0.4 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.1 | 0.3 | GO:0031509 | telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.1 | 0.7 | GO:0014832 | urinary bladder smooth muscle contraction(GO:0014832) |
| 0.1 | 0.3 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.1 | 0.3 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.1 | 0.3 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.1 | 1.5 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 0.5 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.1 | 0.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.1 | 0.7 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.1 | 0.2 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.1 | 0.2 | GO:0051342 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.1 | 0.8 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.1 | 0.2 | GO:0039521 | suppression by virus of host autophagy(GO:0039521) negative regulation of sphingolipid biosynthesis involved in cellular sphingolipid homeostasis(GO:0090157) |
| 0.1 | 0.3 | GO:0006780 | uroporphyrinogen III biosynthetic process(GO:0006780) |
| 0.1 | 0.3 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 0.6 | GO:0045056 | transcytosis(GO:0045056) |
| 0.1 | 0.4 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 1.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.2 | GO:0015960 | diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) |
| 0.1 | 0.2 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.1 | 0.1 | GO:0009227 | nucleotide-sugar catabolic process(GO:0009227) |
| 0.1 | 1.1 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.1 | 1.1 | GO:0046855 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.1 | 0.4 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 0.8 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.1 | 0.2 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.1 | 0.3 | GO:0018002 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.1 | 0.3 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 | 0.2 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.1 | 0.9 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 0.2 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.1 | 0.3 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 0.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 0.6 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.1 | 0.4 | GO:0031282 | regulation of guanylate cyclase activity(GO:0031282) |
| 0.0 | 1.1 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.2 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.6 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.7 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.0 | 0.5 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.0 | 0.6 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.3 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 1.1 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.6 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 | 0.5 | GO:0071360 | cellular response to exogenous dsRNA(GO:0071360) |
| 0.0 | 0.1 | GO:1905005 | regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905005) |
| 0.0 | 1.0 | GO:0001783 | B cell apoptotic process(GO:0001783) |
| 0.0 | 0.2 | GO:0046874 | quinolinate metabolic process(GO:0046874) pyridine-containing compound catabolic process(GO:0072526) |
| 0.0 | 0.5 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) |
| 0.0 | 0.3 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.3 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.0 | 0.2 | GO:0003105 | negative regulation of glomerular filtration(GO:0003105) |
| 0.0 | 0.8 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.2 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.2 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.8 | GO:0035036 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) |
| 0.0 | 0.5 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.0 | 0.6 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 0.2 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.7 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.0 | 2.7 | GO:0001824 | blastocyst development(GO:0001824) |
| 0.0 | 0.2 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.1 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.0 | 0.6 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.2 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.0 | 0.2 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.1 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.0 | 0.7 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.5 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.1 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.0 | 1.4 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.9 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.5 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.0 | 0.4 | GO:0048070 | regulation of developmental pigmentation(GO:0048070) |
| 0.0 | 0.3 | GO:0003094 | glomerular filtration(GO:0003094) renal filtration(GO:0097205) |
| 0.0 | 0.3 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.9 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.7 | GO:0051058 | negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.0 | 0.5 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.3 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.0 | 0.7 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.3 | GO:0001710 | mesodermal cell fate commitment(GO:0001710) |
| 0.0 | 1.6 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.6 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.5 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.1 | GO:0032275 | luteinizing hormone secretion(GO:0032275) positive regulation of gonadotropin secretion(GO:0032278) |
| 0.0 | 0.7 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.8 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.2 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.1 | GO:1903772 | regulation of viral budding via host ESCRT complex(GO:1903772) |
| 0.0 | 0.2 | GO:0032438 | melanosome organization(GO:0032438) pigment granule organization(GO:0048753) |
| 0.0 | 1.4 | GO:0009566 | fertilization(GO:0009566) |
| 0.0 | 0.3 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.7 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.4 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 0.1 | GO:0010726 | positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.0 | 0.2 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.3 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.8 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.2 | GO:0097050 | type B pancreatic cell apoptotic process(GO:0097050) |
| 0.0 | 0.5 | GO:0030819 | positive regulation of cAMP biosynthetic process(GO:0030819) |
| 0.0 | 0.3 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 0.6 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.2 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.0 | 0.0 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.0 | 0.0 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.5 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0000802 | transverse filament(GO:0000802) |
| 0.5 | 1.5 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.3 | 1.5 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.3 | 1.1 | GO:0031230 | intrinsic component of cell outer membrane(GO:0031230) integral component of cell outer membrane(GO:0045203) |
| 0.2 | 0.7 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.2 | 0.9 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.2 | 0.8 | GO:0002177 | manchette(GO:0002177) |
| 0.2 | 0.6 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.1 | 1.8 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 0.4 | GO:1990879 | CST complex(GO:1990879) |
| 0.1 | 1.1 | GO:0090576 | RNA polymerase III transcription factor complex(GO:0090576) |
| 0.1 | 0.7 | GO:0089701 | U2AF(GO:0089701) |
| 0.1 | 1.3 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 1.6 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.1 | 2.7 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 0.8 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.1 | 0.5 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 0.3 | GO:0071010 | prespliceosome(GO:0071010) |
| 0.1 | 1.3 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.1 | 0.3 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.1 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 0.3 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 3.4 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 0.4 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.8 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.1 | 0.7 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 1.3 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 0.4 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 0.2 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.1 | 0.7 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 0.3 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.1 | 0.2 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.3 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.0 | 0.3 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.5 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.6 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.7 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.4 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 1.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 1.0 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.2 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.6 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.6 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.6 | GO:0033202 | DNA helicase complex(GO:0033202) |
| 0.0 | 0.6 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.5 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.2 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.9 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 1.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.8 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.3 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.5 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.5 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.9 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 2.9 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.6 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 1.2 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.3 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 0.0 | 0.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.3 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 1.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.5 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 1.3 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.1 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.2 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.9 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 1.5 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.7 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.9 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.4 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.9 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.6 | 3.9 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.6 | 3.4 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.5 | 1.9 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.5 | 1.4 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.5 | 1.4 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.4 | 2.0 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.3 | 1.0 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) |
| 0.3 | 0.9 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 0.3 | 0.9 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.3 | 1.2 | GO:0042834 | peptidoglycan binding(GO:0042834) CARD domain binding(GO:0050700) |
| 0.3 | 1.4 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.3 | 1.3 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.2 | 0.7 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.2 | 0.9 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.2 | 0.7 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.2 | 1.8 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.2 | 0.5 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.2 | 1.9 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.2 | 0.7 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.2 | 0.6 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.2 | 1.3 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.2 | 2.4 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 0.9 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 1.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 1.0 | GO:0016937 | short-branched-chain-acyl-CoA dehydrogenase activity(GO:0016937) |
| 0.1 | 0.7 | GO:0008761 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) |
| 0.1 | 1.4 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.1 | 2.7 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.1 | 0.4 | GO:0030249 | guanylate cyclase regulator activity(GO:0030249) |
| 0.1 | 0.6 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.1 | 1.1 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.1 | 0.3 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 0.7 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.1 | 2.1 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.1 | 0.2 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.1 | 0.4 | GO:0071532 | ornithine decarboxylase inhibitor activity(GO:0008073) ankyrin repeat binding(GO:0071532) |
| 0.1 | 0.8 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 0.8 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 0.4 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) |
| 0.1 | 0.9 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.1 | 0.3 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.1 | 0.3 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.1 | 3.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.5 | GO:0052630 | CTP:2,3-di-O-geranylgeranyl-sn-glycero-1-phosphate cytidyltransferase activity(GO:0043338) phospholactate guanylyltransferase activity(GO:0043814) ATP:coenzyme F420 adenylyltransferase activity(GO:0043910) UDP-N-acetylgalactosamine diphosphorylase activity(GO:0052630) |
| 0.1 | 0.5 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 1.1 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.1 | 0.4 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.1 | 0.3 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.1 | 0.6 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.1 | 1.5 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 0.2 | GO:0036004 | GAF domain binding(GO:0036004) |
| 0.1 | 0.2 | GO:0019779 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.1 | 1.5 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 0.5 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.1 | 0.2 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 1.2 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 1.2 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 0.3 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.1 | 0.9 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.1 | 0.4 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 0.3 | GO:0004661 | protein geranylgeranyltransferase activity(GO:0004661) |
| 0.1 | 1.1 | GO:0017161 | phosphohistidine phosphatase activity(GO:0008969) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) NADP phosphatase activity(GO:0019178) 5-amino-6-(5-phosphoribitylamino)uracil phosphatase activity(GO:0043726) phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) phosphatidylinositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052867) IDP phosphatase activity(GO:1990003) |
| 0.1 | 0.2 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) |
| 0.1 | 0.4 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.3 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.0 | 0.3 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 2.7 | GO:0035496 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) O antigen polymerase activity(GO:0008755) lipopolysaccharide-1,6-galactosyltransferase activity(GO:0008921) cellulose synthase activity(GO:0016759) 9-phenanthrol UDP-glucuronosyltransferase activity(GO:0018715) 1-phenanthrol glycosyltransferase activity(GO:0018716) 9-phenanthrol glycosyltransferase activity(GO:0018717) 1,2-dihydroxy-phenanthrene glycosyltransferase activity(GO:0018718) phenanthrol glycosyltransferase activity(GO:0019112) alpha-1,2-galactosyltransferase activity(GO:0031278) dolichyl pyrophosphate Man7GlcNAc2 alpha-1,3-glucosyltransferase activity(GO:0033556) endogalactosaminidase activity(GO:0033931) lipopolysaccharide-1,5-galactosyltransferase activity(GO:0035496) dolichyl pyrophosphate Glc1Man9GlcNAc2 alpha-1,3-glucosyltransferase activity(GO:0042283) inositol phosphoceramide synthase activity(GO:0045140) alpha-(1->3)-fucosyltransferase activity(GO:0046920) indole-3-butyrate beta-glucosyltransferase activity(GO:0052638) salicylic acid glucosyltransferase (ester-forming) activity(GO:0052639) salicylic acid glucosyltransferase (glucoside-forming) activity(GO:0052640) benzoic acid glucosyltransferase activity(GO:0052641) chondroitin hydrolase activity(GO:0052757) dolichyl-pyrophosphate Man7GlcNAc2 alpha-1,6-mannosyltransferase activity(GO:0052824) cytokinin 9-beta-glucosyltransferase activity(GO:0080062) |
| 0.0 | 0.8 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.9 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.6 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.4 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.6 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.8 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.4 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.4 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.4 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.1 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 0.3 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 1.5 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.7 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 1.3 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.8 | GO:0034945 | dihydrolipoamide branched chain acyltransferase activity(GO:0004147) sterol O-acyltransferase activity(GO:0004772) palmitoleoyl [acyl-carrier-protein]-dependent acyltransferase activity(GO:0008951) serine O-acyltransferase activity(GO:0016412) O-succinyltransferase activity(GO:0016750) sinapoyltransferase activity(GO:0016752) O-sinapoyltransferase activity(GO:0016753) peptidyl-lysine N6-myristoyltransferase activity(GO:0018030) peptidyl-lysine N6-palmitoyltransferase activity(GO:0018031) benzoyl acetate-CoA thiolase activity(GO:0018711) 3-hydroxybutyryl-CoA thiolase activity(GO:0018712) 3-ketopimelyl-CoA thiolase activity(GO:0018713) N-palmitoyltransferase activity(GO:0019105) acyl-CoA N-acyltransferase activity(GO:0019186) protein-cysteine S-myristoyltransferase activity(GO:0019705) glucosaminyl-phosphotidylinositol O-acyltransferase activity(GO:0032216) ergosterol O-acyltransferase activity(GO:0034737) lanosterol O-acyltransferase activity(GO:0034738) naphthyl-2-oxomethyl-succinyl-CoA succinyl transferase activity(GO:0034848) 2,4,4-trimethyl-3-oxopentanoyl-CoA 2-C-propanoyl transferase activity(GO:0034851) 2-methylhexanoyl-CoA C-acetyltransferase activity(GO:0034915) butyryl-CoA 2-C-propionyltransferase activity(GO:0034919) 2,6-dimethyl-5-methylene-3-oxo-heptanoyl-CoA C-acetyltransferase activity(GO:0034945) L-2-aminoadipate N-acetyltransferase activity(GO:0043741) keto acid formate lyase activity(GO:0043806) azetidine-2-carboxylic acid acetyltransferase activity(GO:0046941) peptidyl-lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0052858) acetyl-CoA:L-lysine N6-acetyltransferase(GO:0090595) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.7 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.5 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.2 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.9 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.2 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) glycerol channel activity(GO:0015254) |
| 0.0 | 0.9 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.4 | GO:0035380 | C-3 sterol dehydrogenase (C-4 sterol decarboxylase) activity(GO:0000252) mevaldate reductase activity(GO:0004495) gluconate dehydrogenase activity(GO:0008875) epoxide dehydrogenase activity(GO:0018451) 5-exo-hydroxycamphor dehydrogenase activity(GO:0018452) 2-hydroxytetrahydrofuran dehydrogenase activity(GO:0018453) acetoin dehydrogenase activity(GO:0019152) phenylcoumaran benzylic ether reductase activity(GO:0032442) D-xylose:NADP reductase activity(GO:0032866) L-arabinose:NADP reductase activity(GO:0032867) D-arabinitol dehydrogenase, D-ribulose forming (NADP+) activity(GO:0033709) (R)-(-)-1,2,3,4-tetrahydronaphthol dehydrogenase activity(GO:0034831) 3-hydroxymenthone dehydrogenase activity(GO:0034840) very long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0035380) dihydrotestosterone 17-beta-dehydrogenase activity(GO:0035410) (R)-2-hydroxyisocaproate dehydrogenase activity(GO:0043713) L-arabinose 1-dehydrogenase (NADP+) activity(GO:0044103) L-xylulose reductase (NAD+) activity(GO:0044105) 3-ketoglucose-reductase activity(GO:0048258) D-arabinitol dehydrogenase, D-xylulose forming (NADP+) activity(GO:0052677) |
| 0.0 | 1.0 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 1.1 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 1.4 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 1.7 | GO:0043851 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) rRNA (uridine-2'-O-)-methyltransferase activity(GO:0008650) rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) selenocysteine methyltransferase activity(GO:0016205) rRNA (adenine) methyltransferase activity(GO:0016433) rRNA (cytosine) methyltransferase activity(GO:0016434) rRNA (guanine) methyltransferase activity(GO:0016435) 1-phenanthrol methyltransferase activity(GO:0018707) protein-arginine N5-methyltransferase activity(GO:0019702) dimethylarsinite methyltransferase activity(GO:0034541) 4,5-dihydroxybenzo(a)pyrene methyltransferase activity(GO:0034807) 1-hydroxypyrene methyltransferase activity(GO:0034931) 1-hydroxy-6-methoxypyrene methyltransferase activity(GO:0034933) demethylmenaquinone methyltransferase activity(GO:0043770) cobalt-precorrin-6B C5-methyltransferase activity(GO:0043776) cobalt-precorrin-7 C15-methyltransferase activity(GO:0043777) cobalt-precorrin-5B C1-methyltransferase activity(GO:0043780) cobalt-precorrin-3 C17-methyltransferase activity(GO:0043782) dimethylamine methyltransferase activity(GO:0043791) hydroxyneurosporene-O-methyltransferase activity(GO:0043803) tRNA (adenine-57, 58-N(1)-) methyltransferase activity(GO:0043827) methylamine-specific methylcobalamin:coenzyme M methyltransferase activity(GO:0043833) trimethylamine methyltransferase activity(GO:0043834) methanol-specific methylcobalamin:coenzyme M methyltransferase activity(GO:0043851) monomethylamine methyltransferase activity(GO:0043852) P-methyltransferase activity(GO:0051994) Se-methyltransferase activity(GO:0051995) 2-phytyl-1,4-naphthoquinone methyltransferase activity(GO:0052624) tRNA (uracil-2'-O-)-methyltransferase activity(GO:0052665) tRNA (cytosine-2'-O-)-methyltransferase activity(GO:0052666) phosphomethylethanolamine N-methyltransferase activity(GO:0052667) tRNA (cytosine-3-)-methyltransferase activity(GO:0052735) rRNA (cytosine-2'-O-)-methyltransferase activity(GO:0070677) rRNA (cytosine-N4-)-methyltransferase activity(GO:0071424) trihydroxyferuloyl spermidine O-methyltransferase activity(GO:0080012) |
| 0.0 | 0.8 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 1.2 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 1.8 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 1.1 | GO:0008186 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 0.1 | GO:0008251 | tRNA-specific adenosine deaminase activity(GO:0008251) |
| 0.0 | 0.5 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 0.3 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 2.5 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 1.9 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 3.8 | GO:0043774 | UDP-N-acetylmuramoylalanyl-D-glutamyl-2,6-diaminopimelate-D-alanyl-D-alanine ligase activity(GO:0008766) ribosomal S6-glutamic acid ligase activity(GO:0018169) coenzyme F420-0 gamma-glutamyl ligase activity(GO:0043773) coenzyme F420-2 alpha-glutamyl ligase activity(GO:0043774) protein-glycine ligase activity(GO:0070735) protein-glycine ligase activity, initiating(GO:0070736) protein-glycine ligase activity, elongating(GO:0070737) tubulin-glycine ligase activity(GO:0070738) |
| 0.0 | 0.2 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.0 | GO:0048039 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) ubiquinone binding(GO:0048039) |
| 0.0 | 0.2 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 2.3 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.1 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.5 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.1 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.3 | GO:0019239 | deaminase activity(GO:0019239) |
| 0.0 | 1.9 | GO:0003729 | mRNA binding(GO:0003729) |
| 0.0 | 0.6 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.5 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |