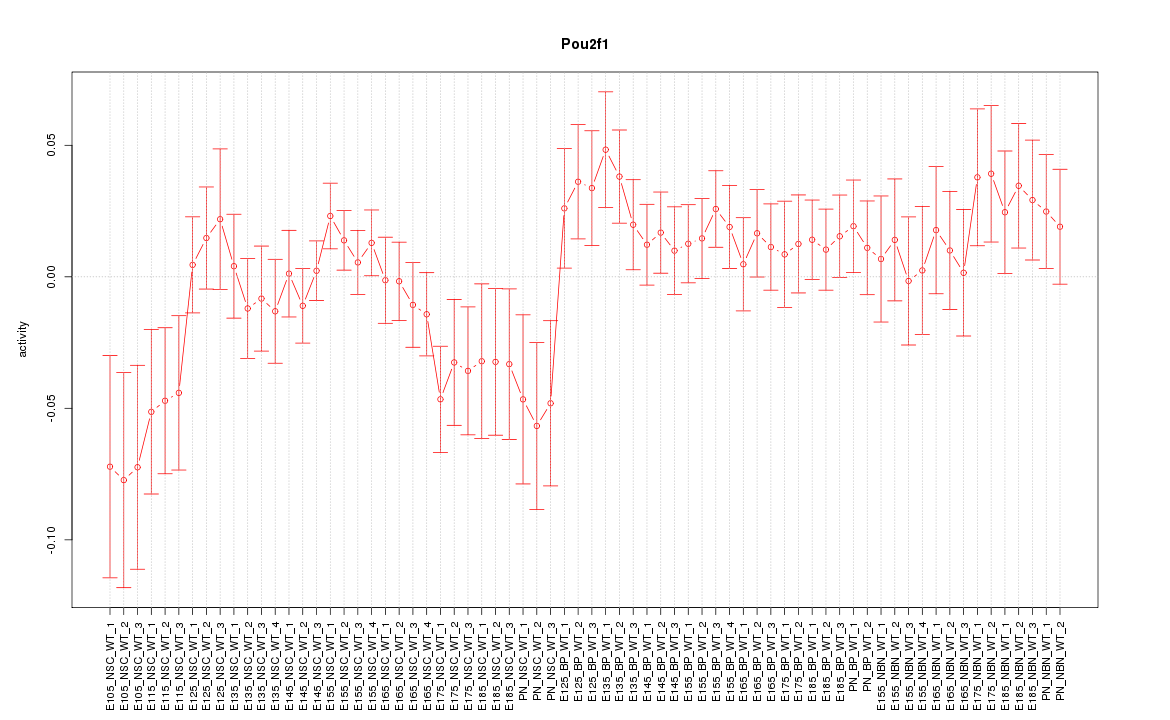
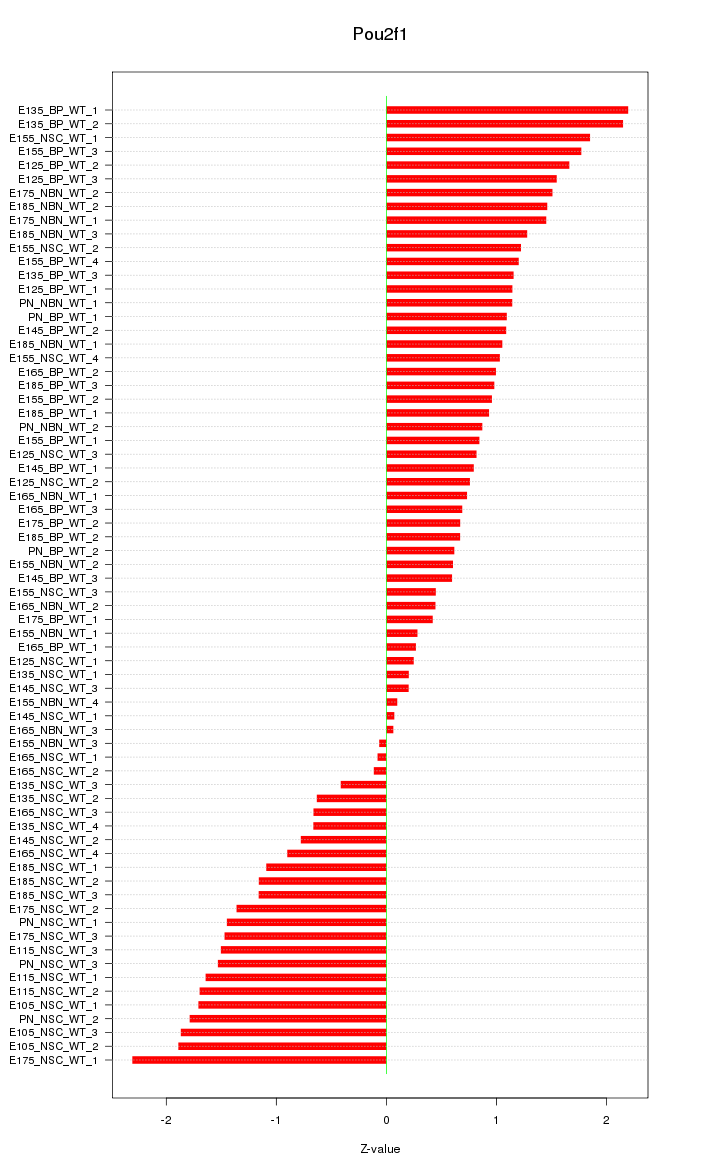
Motif ID: Pou2f1
Z-value: 1.155
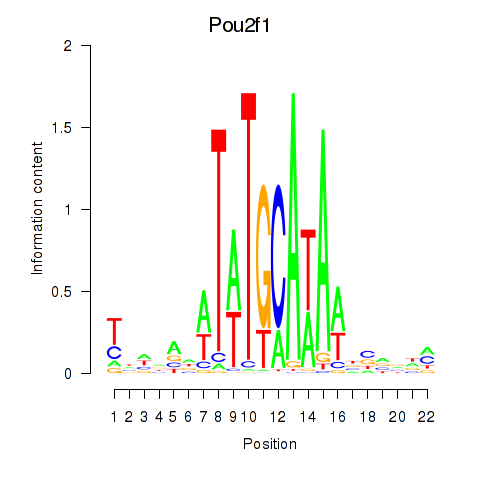
Transcription factors associated with Pou2f1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Pou2f1 | ENSMUSG00000026565.12 | Pou2f1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pou2f1 | mm10_v2_chr1_-_166002613_166002678 | -0.21 | 8.2e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.3 | 13.0 | GO:0007521 | muscle cell fate determination(GO:0007521) cellular response to parathyroid hormone stimulus(GO:0071374) positive regulation of macrophage apoptotic process(GO:2000111) |
| 3.1 | 18.7 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 2.8 | 8.4 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 2.5 | 7.6 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 1.9 | 5.6 | GO:1902605 | heterotrimeric G-protein complex assembly(GO:1902605) |
| 1.6 | 9.6 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 1.6 | 4.8 | GO:0060067 | cervix development(GO:0060067) comma-shaped body morphogenesis(GO:0072049) S-shaped body morphogenesis(GO:0072050) anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 1.4 | 4.3 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 1.3 | 7.8 | GO:0035743 | CD4-positive, alpha-beta T cell cytokine production(GO:0035743) T-helper 2 cell cytokine production(GO:0035745) |
| 1.3 | 2.6 | GO:0060025 | regulation of synaptic activity(GO:0060025) |
| 0.9 | 7.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.9 | 2.6 | GO:0051464 | positive regulation of cortisol secretion(GO:0051464) |
| 0.8 | 4.2 | GO:0010359 | regulation of anion channel activity(GO:0010359) |
| 0.8 | 16.0 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.8 | 2.4 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.8 | 4.6 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.8 | 2.3 | GO:2001187 | positive regulation of CD8-positive, alpha-beta T cell activation(GO:2001187) |
| 0.7 | 3.4 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.6 | 1.9 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.6 | 3.4 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.5 | 6.5 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.5 | 3.1 | GO:0002681 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.5 | 1.5 | GO:1904742 | protein poly-ADP-ribosylation(GO:0070212) regulation of telomeric DNA binding(GO:1904742) |
| 0.5 | 1.5 | GO:0030210 | heparin biosynthetic process(GO:0030210) Tie signaling pathway(GO:0048014) |
| 0.5 | 1.9 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.5 | 1.8 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.4 | 1.8 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.4 | 3.6 | GO:1901030 | positive regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901030) |
| 0.4 | 5.6 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.4 | 1.1 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.4 | 6.8 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.3 | 2.8 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.3 | 6.4 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.3 | 2.4 | GO:1901162 | tryptophan metabolic process(GO:0006568) indolalkylamine metabolic process(GO:0006586) serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.3 | 3.1 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.3 | 6.8 | GO:1902850 | mitotic spindle assembly(GO:0090307) microtubule cytoskeleton organization involved in mitosis(GO:1902850) |
| 0.3 | 3.6 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.3 | 0.8 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.3 | 4.9 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.3 | 26.7 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.2 | 6.2 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 0.7 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 0.2 | 2.9 | GO:0036120 | cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.2 | 0.9 | GO:0072592 | regulation of integrin biosynthetic process(GO:0045113) oxygen metabolic process(GO:0072592) |
| 0.2 | 7.1 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.2 | 0.6 | GO:0019478 | D-amino acid catabolic process(GO:0019478) |
| 0.2 | 27.4 | GO:0006333 | chromatin assembly or disassembly(GO:0006333) |
| 0.2 | 0.7 | GO:0043461 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.2 | 0.5 | GO:0034310 | primary alcohol catabolic process(GO:0034310) |
| 0.2 | 9.8 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.2 | 3.6 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.2 | 1.0 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.1 | 0.9 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 3.1 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.1 | 2.6 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.1 | 0.8 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.1 | 8.4 | GO:0008542 | visual learning(GO:0008542) |
| 0.1 | 9.0 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 0.7 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 2.4 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.1 | 2.3 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 7.5 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 6.0 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.1 | 0.5 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 0.1 | 0.7 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.1 | 0.9 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.1 | 0.6 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.1 | 1.2 | GO:0032094 | response to food(GO:0032094) |
| 0.1 | 3.1 | GO:0032410 | negative regulation of transporter activity(GO:0032410) |
| 0.1 | 1.6 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 1.2 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.3 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.0 | 0.5 | GO:0030157 | pancreatic juice secretion(GO:0030157) |
| 0.0 | 2.6 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 2.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.9 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 4.8 | GO:0098656 | anion transmembrane transport(GO:0098656) |
| 0.0 | 0.6 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.0 | 0.3 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 1.7 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 2.4 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.0 | 0.4 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 1.6 | GO:0097202 | activation of cysteine-type endopeptidase activity(GO:0097202) |
| 0.0 | 0.9 | GO:0045664 | regulation of neuron differentiation(GO:0045664) |
| 0.0 | 1.3 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 1.4 | GO:0007569 | cell aging(GO:0007569) |
| 0.0 | 0.7 | GO:0006352 | DNA-templated transcription, initiation(GO:0006352) |
| 0.0 | 1.6 | GO:0098792 | xenophagy(GO:0098792) |
| 0.0 | 2.4 | GO:0019827 | stem cell population maintenance(GO:0019827) |
| 0.0 | 0.0 | GO:2000343 | positive regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000343) |
| 0.0 | 0.2 | GO:0044406 | adhesion of symbiont to host(GO:0044406) |
| 0.0 | 0.4 | GO:0002181 | cytoplasmic translation(GO:0002181) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.4 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 1.1 | 6.8 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.9 | 17.5 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.6 | 3.1 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.6 | 5.7 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.5 | 7.2 | GO:0031045 | dense core granule(GO:0031045) |
| 0.4 | 6.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.3 | 2.4 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.3 | 5.6 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.3 | 1.8 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.2 | 3.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.2 | 1.8 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.2 | 0.9 | GO:0071438 | NADPH oxidase complex(GO:0043020) invadopodium membrane(GO:0071438) |
| 0.2 | 4.2 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.2 | 7.5 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.2 | 0.6 | GO:0070522 | ERCC4-ERCC1 complex(GO:0070522) |
| 0.2 | 6.4 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.2 | 3.8 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.1 | 16.0 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 2.3 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 1.1 | GO:0060091 | kinocilium(GO:0060091) |
| 0.1 | 1.4 | GO:0030867 | desmosome(GO:0030057) rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 3.8 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 13.7 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 0.6 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 2.0 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.1 | 2.4 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 0.8 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 1.6 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 7.9 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 3.6 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 1.0 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.5 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.3 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 3.7 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 2.9 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 2.5 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 1.8 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 26.4 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 1.5 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 1.4 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 3.1 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 0.2 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 1.4 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 1.4 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 2.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.9 | GO:0043235 | receptor complex(GO:0043235) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.6 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 1.9 | 5.6 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 1.2 | 3.6 | GO:0042030 | ATPase inhibitor activity(GO:0042030) angiostatin binding(GO:0043532) |
| 1.2 | 13.0 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.9 | 2.6 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.8 | 4.2 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.8 | 5.6 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.8 | 2.4 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.6 | 2.9 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.5 | 3.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.5 | 3.1 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.5 | 2.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.4 | 8.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.4 | 4.8 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.4 | 1.9 | GO:0042895 | antibiotic transporter activity(GO:0042895) |
| 0.3 | 4.3 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.3 | 2.3 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.3 | 6.4 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.3 | 2.6 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.3 | 2.8 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.3 | 7.0 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.3 | 9.6 | GO:0034930 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) heparan sulfate 2-O-sulfotransferase activity(GO:0004394) HNK-1 sulfotransferase activity(GO:0016232) heparan sulfate 6-O-sulfotransferase activity(GO:0017095) trans-9R,10R-dihydrodiolphenanthrene sulfotransferase activity(GO:0018721) 1-phenanthrol sulfotransferase activity(GO:0018722) 3-phenanthrol sulfotransferase activity(GO:0018723) 4-phenanthrol sulfotransferase activity(GO:0018724) trans-3,4-dihydrodiolphenanthrene sulfotransferase activity(GO:0018725) 9-phenanthrol sulfotransferase activity(GO:0018726) 2-phenanthrol sulfotransferase activity(GO:0018727) phenanthrol sulfotransferase activity(GO:0019111) 1-hydroxypyrene sulfotransferase activity(GO:0034930) proteoglycan sulfotransferase activity(GO:0050698) cholesterol sulfotransferase activity(GO:0051922) hydroxyjasmonate sulfotransferase activity(GO:0080131) |
| 0.3 | 0.9 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.3 | 1.4 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.3 | 5.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.3 | 3.6 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.2 | 1.8 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.2 | 3.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.2 | 8.4 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.2 | 4.0 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.2 | 6.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 0.7 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.1 | 16.0 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.1 | 4.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 2.6 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 1.5 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.1 | 2.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 0.8 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 0.8 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 3.1 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 3.7 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 1.8 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 2.1 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 1.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 2.4 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 4.8 | GO:0008186 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.1 | 0.2 | GO:0019002 | GMP binding(GO:0019002) |
| 0.1 | 5.8 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.1 | 9.8 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 1.7 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 0.5 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 6.5 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.5 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 3.1 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 11.9 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 3.8 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.0 | 3.6 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.3 | GO:0008235 | metalloexopeptidase activity(GO:0008235) |
| 0.0 | 2.6 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 3.6 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 19.8 | GO:0003700 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 0.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 1.1 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.7 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.0 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.4 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 1.6 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.7 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 13.3 | GO:0003677 | DNA binding(GO:0003677) |