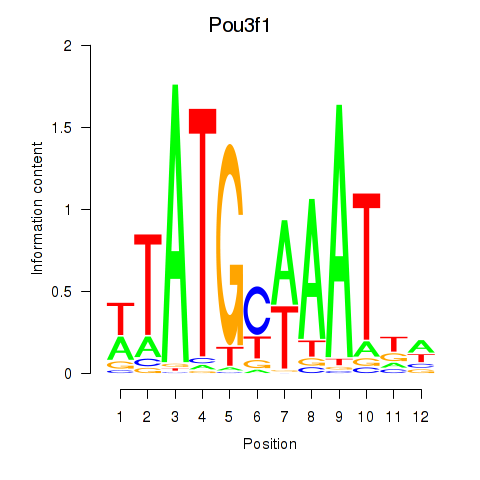
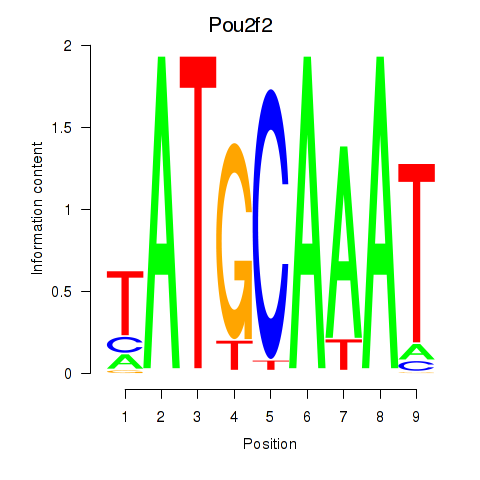
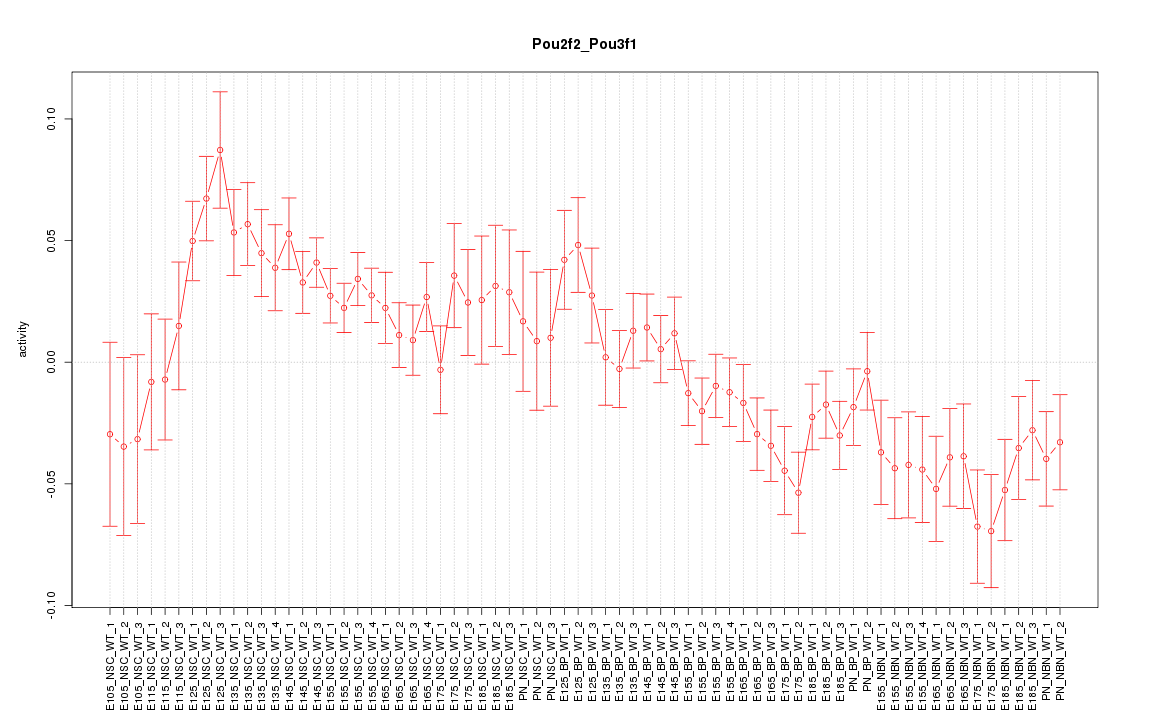
| 5.5 |
27.6 |
GO:0021905 |
pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) |
| 4.8 |
9.6 |
GO:0061344 |
regulation of cell adhesion involved in heart morphogenesis(GO:0061344) positive regulation of ephrin receptor signaling pathway(GO:1901189) |
| 4.5 |
13.5 |
GO:0060853 |
arterial endothelial cell fate commitment(GO:0060844) blood vessel endothelial cell fate commitment(GO:0060846) endothelial cell fate specification(GO:0060847) Notch signaling pathway involved in arterial endothelial cell fate commitment(GO:0060853) blood vessel endothelial cell fate specification(GO:0097101) |
| 4.1 |
12.2 |
GO:0021893 |
cerebral cortex GABAergic interneuron fate commitment(GO:0021893) |
| 3.6 |
10.9 |
GO:0060598 |
dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 3.6 |
10.7 |
GO:1904956 |
dermatome development(GO:0061054) regulation of dermatome development(GO:0061183) regulation of midbrain dopaminergic neuron differentiation(GO:1904956) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 3.1 |
9.3 |
GO:1904059 |
regulation of locomotor rhythm(GO:1904059) |
| 2.6 |
15.8 |
GO:1904936 |
cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 2.4 |
29.4 |
GO:0002227 |
innate immune response in mucosa(GO:0002227) |
| 2.3 |
18.5 |
GO:0014807 |
regulation of somitogenesis(GO:0014807) |
| 2.1 |
27.4 |
GO:0000185 |
activation of MAPKKK activity(GO:0000185) |
| 2.0 |
13.9 |
GO:0031536 |
positive regulation of exit from mitosis(GO:0031536) |
| 2.0 |
5.9 |
GO:0010986 |
positive regulation of lipoprotein particle clearance(GO:0010986) |
| 2.0 |
5.9 |
GO:0060842 |
arterial endothelial cell differentiation(GO:0060842) |
| 2.0 |
29.4 |
GO:0034501 |
protein localization to kinetochore(GO:0034501) |
| 1.8 |
3.5 |
GO:0021759 |
globus pallidus development(GO:0021759) |
| 1.8 |
5.3 |
GO:0006553 |
lysine metabolic process(GO:0006553) |
| 1.4 |
4.2 |
GO:1901896 |
positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 1.3 |
7.6 |
GO:0002326 |
B cell lineage commitment(GO:0002326) |
| 1.3 |
3.8 |
GO:0070269 |
pyroptosis(GO:0070269) |
| 1.2 |
3.7 |
GO:1903998 |
regulation of eating behavior(GO:1903998) |
| 1.2 |
19.8 |
GO:0033234 |
negative regulation of protein sumoylation(GO:0033234) |
| 1.1 |
16.8 |
GO:0035563 |
positive regulation of chromatin binding(GO:0035563) |
| 1.1 |
4.4 |
GO:0046552 |
eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 1.1 |
4.3 |
GO:0043973 |
histone H3-K4 acetylation(GO:0043973) |
| 1.1 |
15.1 |
GO:0090129 |
positive regulation of synapse maturation(GO:0090129) |
| 1.0 |
4.1 |
GO:0014012 |
peripheral nervous system axon regeneration(GO:0014012) |
| 1.0 |
3.1 |
GO:0090425 |
hepatocyte cell migration(GO:0002194) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 1.0 |
7.9 |
GO:0018401 |
peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.9 |
8.3 |
GO:0048664 |
neuron fate determination(GO:0048664) |
| 0.9 |
6.2 |
GO:0000320 |
re-entry into mitotic cell cycle(GO:0000320) |
| 0.9 |
11.5 |
GO:0021521 |
ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.8 |
25.2 |
GO:0007638 |
mechanosensory behavior(GO:0007638) |
| 0.8 |
11.9 |
GO:0097237 |
cellular response to toxic substance(GO:0097237) |
| 0.8 |
2.3 |
GO:0021546 |
rhombomere development(GO:0021546) |
| 0.7 |
3.0 |
GO:0090324 |
negative regulation of oxidative phosphorylation(GO:0090324) |
| 0.7 |
78.5 |
GO:0006342 |
chromatin silencing(GO:0006342) |
| 0.7 |
1.4 |
GO:0043490 |
malate-aspartate shuttle(GO:0043490) |
| 0.7 |
5.6 |
GO:0097341 |
inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.7 |
4.9 |
GO:1903715 |
regulation of aerobic respiration(GO:1903715) |
| 0.7 |
2.0 |
GO:0016332 |
establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.6 |
9.6 |
GO:0021891 |
olfactory bulb interneuron development(GO:0021891) |
| 0.6 |
5.0 |
GO:0006265 |
DNA topological change(GO:0006265) |
| 0.6 |
27.3 |
GO:0060325 |
face morphogenesis(GO:0060325) |
| 0.6 |
6.2 |
GO:1902231 |
positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.6 |
2.3 |
GO:0090086 |
negative regulation of protein deubiquitination(GO:0090086) |
| 0.6 |
1.7 |
GO:2000054 |
regulation of centromeric sister chromatid cohesion(GO:0070602) regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.5 |
3.3 |
GO:0072513 |
positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.5 |
1.6 |
GO:2000065 |
negative regulation of aldosterone metabolic process(GO:0032345) negative regulation of aldosterone biosynthetic process(GO:0032348) regulation of cortisol biosynthetic process(GO:2000064) negative regulation of cortisol biosynthetic process(GO:2000065) |
| 0.5 |
4.3 |
GO:0034983 |
peptidyl-lysine deacetylation(GO:0034983) regulation of skeletal muscle fiber development(GO:0048742) histone H4 deacetylation(GO:0070933) |
| 0.5 |
7.1 |
GO:0060074 |
synapse maturation(GO:0060074) |
| 0.5 |
17.7 |
GO:0000038 |
very long-chain fatty acid metabolic process(GO:0000038) |
| 0.5 |
2.3 |
GO:2000255 |
negative regulation of male germ cell proliferation(GO:2000255) |
| 0.4 |
9.7 |
GO:0001967 |
suckling behavior(GO:0001967) |
| 0.4 |
0.8 |
GO:0090085 |
regulation of protein deubiquitination(GO:0090085) |
| 0.4 |
5.9 |
GO:0048387 |
negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.4 |
2.2 |
GO:0002072 |
optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.4 |
2.8 |
GO:0048733 |
sebaceous gland development(GO:0048733) |
| 0.3 |
1.5 |
GO:0035093 |
spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.3 |
2.4 |
GO:0048023 |
positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.3 |
28.5 |
GO:0006334 |
nucleosome assembly(GO:0006334) |
| 0.3 |
0.8 |
GO:0001978 |
regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) |
| 0.3 |
11.3 |
GO:0006284 |
base-excision repair(GO:0006284) |
| 0.2 |
2.9 |
GO:1904262 |
negative regulation of TORC1 signaling(GO:1904262) |
| 0.2 |
4.1 |
GO:0010569 |
regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.2 |
2.3 |
GO:0060736 |
prostate gland growth(GO:0060736) |
| 0.2 |
1.3 |
GO:0090209 |
negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.2 |
4.6 |
GO:0070306 |
lens fiber cell differentiation(GO:0070306) |
| 0.2 |
3.4 |
GO:2000001 |
regulation of DNA damage checkpoint(GO:2000001) |
| 0.2 |
1.2 |
GO:2000210 |
positive regulation of anoikis(GO:2000210) |
| 0.2 |
1.6 |
GO:0006477 |
protein sulfation(GO:0006477) |
| 0.2 |
0.6 |
GO:0045014 |
detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) detection of glucose(GO:0051594) |
| 0.2 |
1.3 |
GO:1904217 |
regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.2 |
1.3 |
GO:0070561 |
vitamin D receptor signaling pathway(GO:0070561) |
| 0.2 |
7.7 |
GO:0043029 |
T cell homeostasis(GO:0043029) |
| 0.2 |
5.2 |
GO:0003301 |
physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.2 |
0.3 |
GO:0060385 |
axonogenesis involved in innervation(GO:0060385) |
| 0.2 |
6.5 |
GO:0032611 |
interleukin-1 beta production(GO:0032611) |
| 0.1 |
0.6 |
GO:0014846 |
esophagus smooth muscle contraction(GO:0014846) |
| 0.1 |
1.3 |
GO:0006268 |
DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 |
2.2 |
GO:0050860 |
negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 |
6.0 |
GO:0035307 |
positive regulation of protein dephosphorylation(GO:0035307) |
| 0.1 |
7.0 |
GO:0002066 |
columnar/cuboidal epithelial cell development(GO:0002066) |
| 0.1 |
2.8 |
GO:0044839 |
G2/M transition of mitotic cell cycle(GO:0000086) cell cycle G2/M phase transition(GO:0044839) |
| 0.1 |
0.7 |
GO:2000124 |
regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 |
2.5 |
GO:0021884 |
forebrain neuron development(GO:0021884) |
| 0.1 |
0.8 |
GO:0002082 |
regulation of oxidative phosphorylation(GO:0002082) |
| 0.1 |
4.9 |
GO:0021954 |
central nervous system neuron development(GO:0021954) |
| 0.1 |
1.7 |
GO:0016180 |
snRNA processing(GO:0016180) |
| 0.1 |
0.8 |
GO:0060065 |
uterus development(GO:0060065) |
| 0.1 |
2.3 |
GO:0036075 |
endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 |
1.0 |
GO:0002115 |
store-operated calcium entry(GO:0002115) |
| 0.1 |
1.9 |
GO:0010842 |
retina layer formation(GO:0010842) |
| 0.1 |
1.2 |
GO:0046069 |
cGMP catabolic process(GO:0046069) |
| 0.1 |
0.6 |
GO:2000381 |
negative regulation of mesoderm development(GO:2000381) |
| 0.1 |
1.7 |
GO:0030513 |
positive regulation of BMP signaling pathway(GO:0030513) |
| 0.1 |
0.2 |
GO:0060235 |
lens induction in camera-type eye(GO:0060235) |
| 0.1 |
1.9 |
GO:0035019 |
somatic stem cell population maintenance(GO:0035019) |
| 0.1 |
2.3 |
GO:0045670 |
regulation of osteoclast differentiation(GO:0045670) |
| 0.0 |
5.1 |
GO:0010811 |
positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 |
18.0 |
GO:0007067 |
mitotic nuclear division(GO:0007067) |
| 0.0 |
2.3 |
GO:1900046 |
regulation of blood coagulation(GO:0030193) regulation of hemostasis(GO:1900046) |
| 0.0 |
0.4 |
GO:0060056 |
mammary gland involution(GO:0060056) |
| 0.0 |
0.3 |
GO:0060746 |
parental behavior(GO:0060746) |
| 0.0 |
0.2 |
GO:0048149 |
adult feeding behavior(GO:0008343) behavioral response to ethanol(GO:0048149) righting reflex(GO:0060013) |
| 0.0 |
6.0 |
GO:0008360 |
regulation of cell shape(GO:0008360) |
| 0.0 |
2.6 |
GO:0006261 |
DNA-dependent DNA replication(GO:0006261) |
| 0.0 |
0.8 |
GO:0007608 |
sensory perception of smell(GO:0007608) |
| 0.0 |
0.3 |
GO:0006958 |
complement activation, classical pathway(GO:0006958) |
| 0.0 |
3.4 |
GO:0046777 |
protein autophosphorylation(GO:0046777) |
| 0.0 |
1.1 |
GO:0043488 |
regulation of mRNA stability(GO:0043488) |
| 0.0 |
2.8 |
GO:0051260 |
protein homooligomerization(GO:0051260) |
| 0.0 |
0.3 |
GO:0030336 |
negative regulation of cell migration(GO:0030336) |