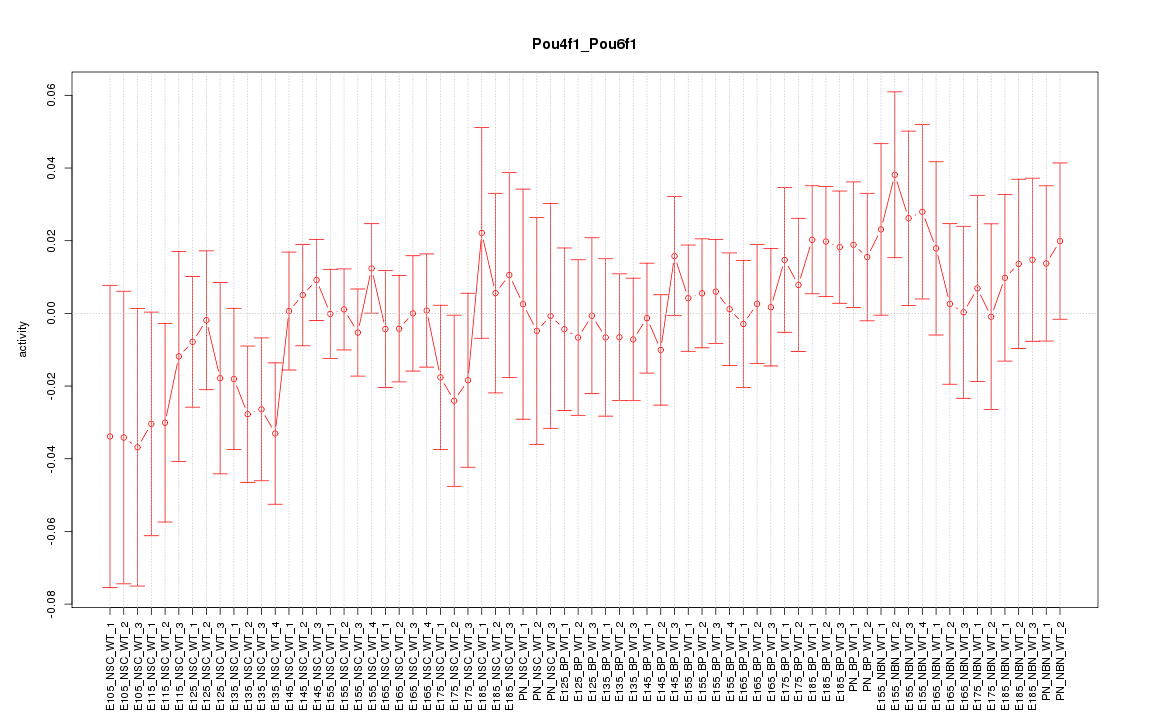
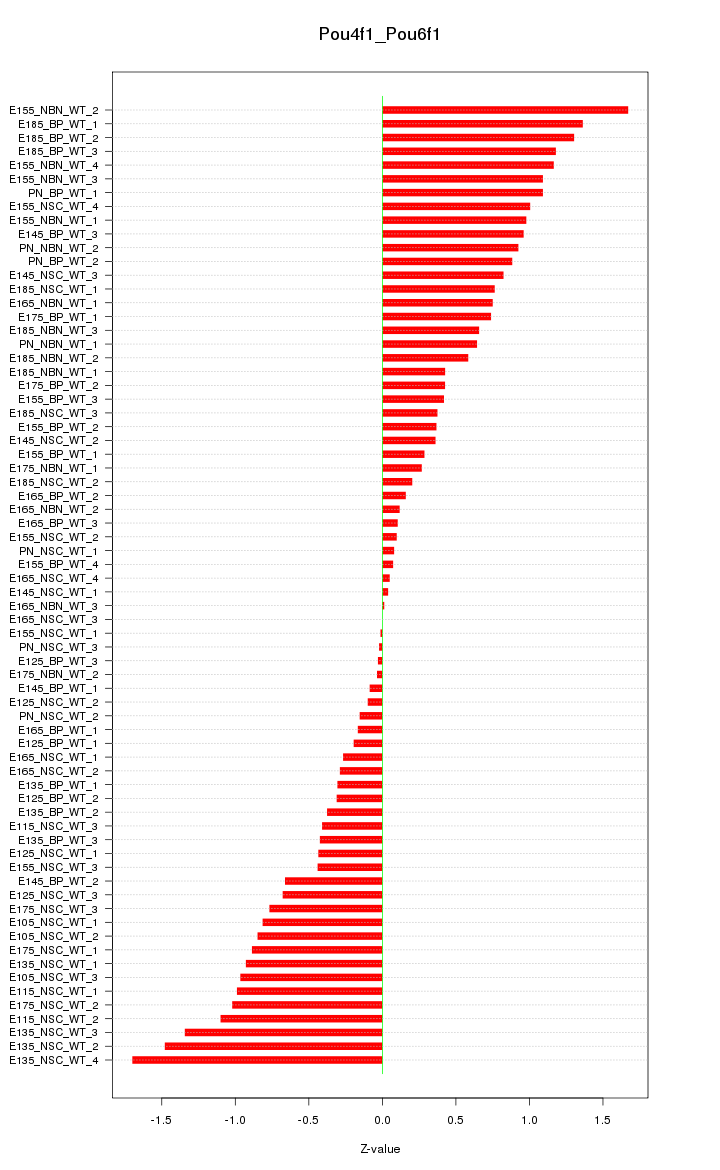
Motif ID: Pou4f1_Pou6f1
Z-value: 0.734
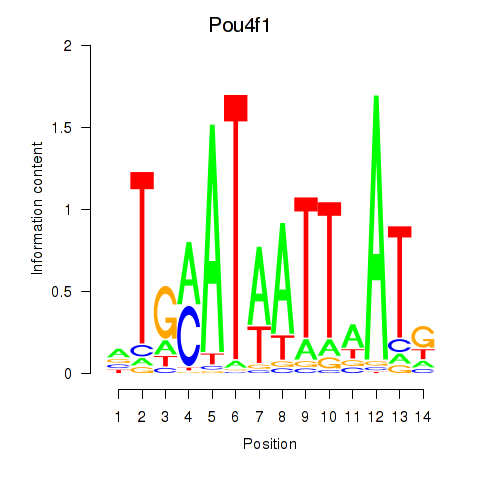
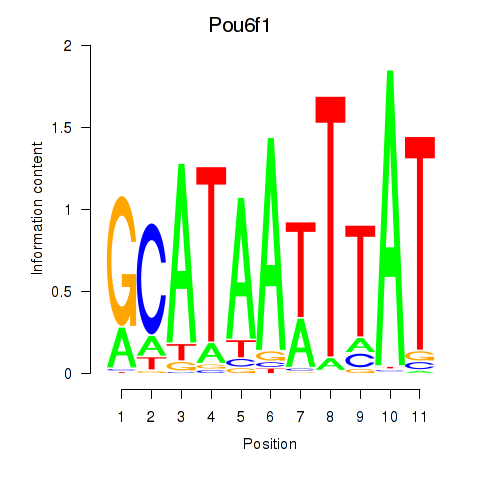
Transcription factors associated with Pou4f1_Pou6f1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Pou4f1 | ENSMUSG00000048349.8 | Pou4f1 |
| Pou6f1 | ENSMUSG00000009739.10 | Pou6f1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pou6f1 | mm10_v2_chr15_-_100599983_100600039 | 0.68 | 1.2e-10 | Click! |
| Pou4f1 | mm10_v2_chr14_-_104467984_104468041 | -0.43 | 2.3e-04 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.3 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 1.6 | 4.8 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 1.1 | 4.3 | GO:0009414 | response to water deprivation(GO:0009414) |
| 1.0 | 2.9 | GO:0007521 | muscle cell fate determination(GO:0007521) cellular response to parathyroid hormone stimulus(GO:0071374) positive regulation of macrophage apoptotic process(GO:2000111) |
| 1.0 | 4.8 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.9 | 5.1 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.8 | 1.5 | GO:0014877 | response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.7 | 8.5 | GO:1901898 | negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.6 | 1.9 | GO:1904395 | regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904393) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.6 | 2.5 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.5 | 2.6 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.5 | 2.6 | GO:0021856 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) facioacoustic ganglion development(GO:1903375) |
| 0.5 | 2.1 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.5 | 1.5 | GO:0042939 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.4 | 2.2 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.4 | 6.2 | GO:1901629 | regulation of presynaptic membrane organization(GO:1901629) |
| 0.4 | 5.7 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.4 | 1.2 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.4 | 1.2 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.4 | 2.3 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.4 | 4.5 | GO:1902993 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.4 | 1.9 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.4 | 0.7 | GO:0031394 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.4 | 5.9 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.3 | 1.6 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.3 | 2.9 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.3 | 2.5 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.3 | 1.0 | GO:0045404 | positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.3 | 7.2 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.3 | 1.1 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 0.3 | 4.0 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.3 | 2.8 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.3 | 0.5 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.3 | 1.8 | GO:0097460 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) ferrous iron import into cell(GO:0097460) |
| 0.3 | 2.0 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.2 | 0.7 | GO:2000564 | CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) positive regulation of CD8-positive, alpha-beta T cell activation(GO:2001187) |
| 0.2 | 1.7 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.2 | 0.5 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.2 | 5.0 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.2 | 1.2 | GO:0035356 | cellular triglyceride homeostasis(GO:0035356) |
| 0.2 | 1.8 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.2 | 12.2 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.2 | 1.0 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.2 | 1.1 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.2 | 2.2 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.2 | 0.5 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.1 | 0.9 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.1 | 0.9 | GO:0043206 | extracellular fibril organization(GO:0043206) |
| 0.1 | 1.8 | GO:0034390 | smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.1 | 2.6 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 0.5 | GO:0006780 | uroporphyrinogen III biosynthetic process(GO:0006780) |
| 0.1 | 1.1 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.1 | 0.9 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 0.7 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.1 | 0.3 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.1 | 0.6 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.1 | 0.3 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.1 | 0.6 | GO:0002681 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.1 | 0.7 | GO:1900194 | negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 0.9 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.1 | 9.2 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.1 | 2.4 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.1 | 0.4 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.1 | 0.3 | GO:0031509 | telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.1 | 1.5 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.1 | 3.0 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 0.5 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.1 | 1.5 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 0.7 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 2.5 | GO:0009395 | phospholipid catabolic process(GO:0009395) |
| 0.1 | 0.3 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.1 | 5.1 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.1 | 0.2 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.1 | 1.3 | GO:0014829 | vascular smooth muscle contraction(GO:0014829) |
| 0.1 | 0.9 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 1.1 | GO:0001553 | luteinization(GO:0001553) |
| 0.1 | 0.7 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 0.4 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.1 | 1.0 | GO:0051926 | negative regulation of calcium ion transport(GO:0051926) |
| 0.1 | 3.7 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.5 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.2 | GO:0036337 | Fas signaling pathway(GO:0036337) |
| 0.0 | 0.6 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 0.7 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 2.1 | GO:0032410 | negative regulation of transporter activity(GO:0032410) |
| 0.0 | 0.5 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 1.0 | GO:0031103 | axon regeneration(GO:0031103) |
| 0.0 | 0.1 | GO:0071139 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) resolution of recombination intermediates(GO:0071139) |
| 0.0 | 0.6 | GO:0032402 | melanosome transport(GO:0032402) |
| 0.0 | 0.6 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.1 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 | 1.0 | GO:0051220 | cytoplasmic sequestering of protein(GO:0051220) |
| 0.0 | 0.6 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.0 | 0.5 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 1.9 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.3 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 2.6 | GO:0050770 | regulation of axonogenesis(GO:0050770) |
| 0.0 | 0.6 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 1.5 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 1.1 | GO:0060349 | bone morphogenesis(GO:0060349) |
| 0.0 | 0.2 | GO:0060746 | parental behavior(GO:0060746) |
| 0.0 | 1.9 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 1.5 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.0 | 0.1 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.8 | GO:0032006 | regulation of TOR signaling(GO:0032006) |
| 0.0 | 1.6 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.1 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.5 | 1.6 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.5 | 5.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.4 | 2.0 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.4 | 2.2 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.3 | 2.2 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.3 | 1.3 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.3 | 8.5 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.2 | 4.4 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.2 | 2.6 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.2 | 0.9 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.2 | 1.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.2 | 5.1 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.2 | 2.4 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.1 | 6.2 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 0.6 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 1.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 0.6 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.1 | 0.5 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 1.9 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 2.6 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 0.7 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.1 | 0.5 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.1 | 2.5 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 1.0 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 0.7 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.1 | 5.0 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 0.7 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 15.0 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 6.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 6.2 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.2 | GO:0097361 | CIA complex(GO:0097361) |
| 0.0 | 3.9 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.6 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.6 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 4.3 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 1.9 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.5 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.4 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 1.5 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 4.2 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.3 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 11.6 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 0.6 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 3.2 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.5 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.6 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.5 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 2.6 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.1 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.1 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.8 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 2.3 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 2.6 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.5 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0031752 | D3 dopamine receptor binding(GO:0031750) D5 dopamine receptor binding(GO:0031752) |
| 0.9 | 5.3 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.7 | 2.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.7 | 2.8 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.6 | 1.9 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.6 | 1.9 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.5 | 2.6 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.5 | 2.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.5 | 2.0 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.5 | 1.8 | GO:0004954 | icosanoid receptor activity(GO:0004953) prostanoid receptor activity(GO:0004954) prostaglandin receptor activity(GO:0004955) |
| 0.4 | 7.6 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.4 | 3.4 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.4 | 2.4 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.4 | 1.2 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.4 | 5.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.3 | 1.0 | GO:0004658 | propionyl-CoA carboxylase activity(GO:0004658) |
| 0.3 | 1.7 | GO:0008808 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) |
| 0.3 | 2.3 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.3 | 2.9 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.3 | 1.6 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.3 | 1.8 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.3 | 5.0 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.3 | 1.8 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.2 | 6.2 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.2 | 4.4 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.2 | 2.6 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.2 | 1.5 | GO:0015651 | quaternary ammonium group transmembrane transporter activity(GO:0015651) |
| 0.2 | 2.5 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.2 | 8.4 | GO:0004120 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) photoreceptor cyclic-nucleotide phosphodiesterase activity(GO:0004120) 7,8-dihydro-D-neopterin 2',3'-cyclic phosphate phosphodiesterase activity(GO:0044688) inositol phosphosphingolipid phospholipase activity(GO:0052712) inositol phosphorylceramide phospholipase activity(GO:0052713) mannosyl-inositol phosphorylceramide phospholipase activity(GO:0052714) mannosyl-diinositol phosphorylceramide phospholipase activity(GO:0052715) |
| 0.2 | 1.2 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.2 | 0.6 | GO:0004008 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.2 | 0.7 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) phospholipase A2 inhibitor activity(GO:0019834) |
| 0.2 | 1.8 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.2 | 0.9 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.2 | 0.9 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.2 | 2.2 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.2 | 0.5 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.2 | 1.0 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.2 | 0.9 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.2 | 0.6 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 1.2 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.1 | 3.0 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 2.5 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 0.8 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 1.7 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.1 | 2.4 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.1 | 0.3 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.1 | 0.5 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 2.3 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 0.5 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.1 | 0.1 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.1 | 4.8 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 0.3 | GO:0050347 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.1 | 0.5 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 2.1 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 5.1 | GO:0001948 | glycoprotein binding(GO:0001948) |
| 0.0 | 3.8 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.3 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 5.8 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 4.0 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 1.2 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.2 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 1.9 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.4 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.2 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 1.9 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 4.9 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.5 | GO:0035380 | C-3 sterol dehydrogenase (C-4 sterol decarboxylase) activity(GO:0000252) mevaldate reductase activity(GO:0004495) gluconate dehydrogenase activity(GO:0008875) epoxide dehydrogenase activity(GO:0018451) 5-exo-hydroxycamphor dehydrogenase activity(GO:0018452) 2-hydroxytetrahydrofuran dehydrogenase activity(GO:0018453) acetoin dehydrogenase activity(GO:0019152) phenylcoumaran benzylic ether reductase activity(GO:0032442) D-xylose:NADP reductase activity(GO:0032866) L-arabinose:NADP reductase activity(GO:0032867) D-arabinitol dehydrogenase, D-ribulose forming (NADP+) activity(GO:0033709) (R)-(-)-1,2,3,4-tetrahydronaphthol dehydrogenase activity(GO:0034831) 3-hydroxymenthone dehydrogenase activity(GO:0034840) very long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0035380) dihydrotestosterone 17-beta-dehydrogenase activity(GO:0035410) (R)-2-hydroxyisocaproate dehydrogenase activity(GO:0043713) L-arabinose 1-dehydrogenase (NADP+) activity(GO:0044103) L-xylulose reductase (NAD+) activity(GO:0044105) 3-ketoglucose-reductase activity(GO:0048258) D-arabinitol dehydrogenase, D-xylulose forming (NADP+) activity(GO:0052677) |
| 0.0 | 0.7 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.6 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.4 | GO:0001098 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.6 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.6 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.3 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 1.2 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.0 | 0.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.5 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 2.7 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 1.6 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.5 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.4 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |