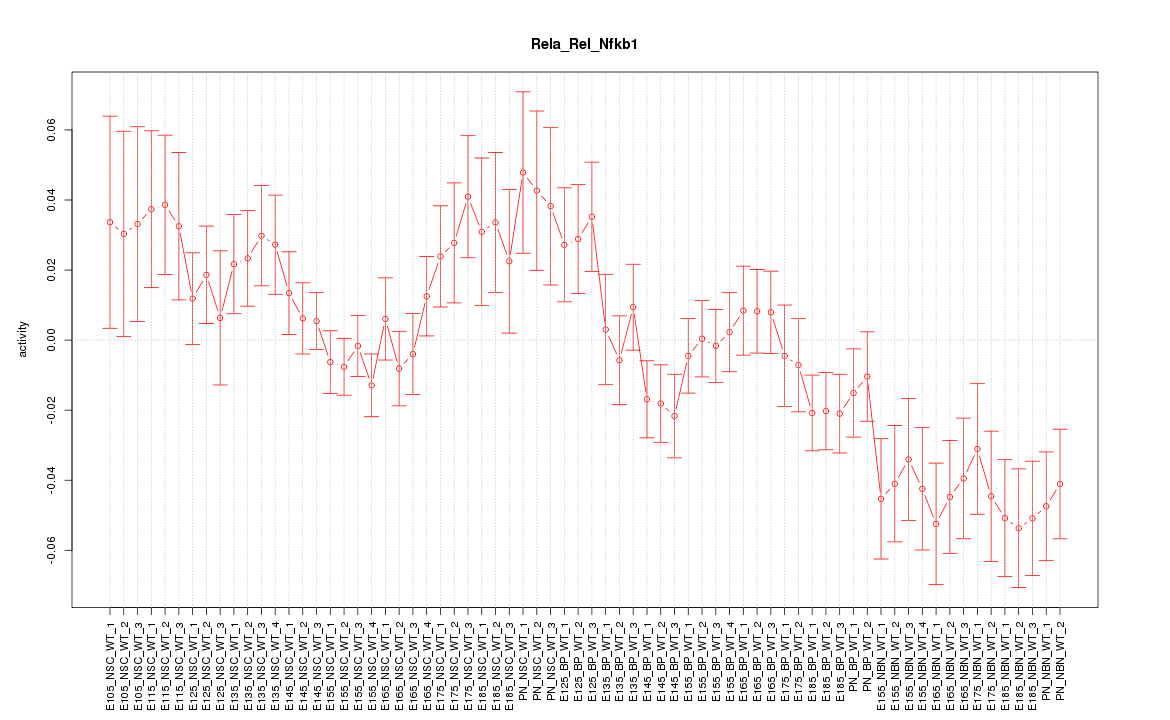
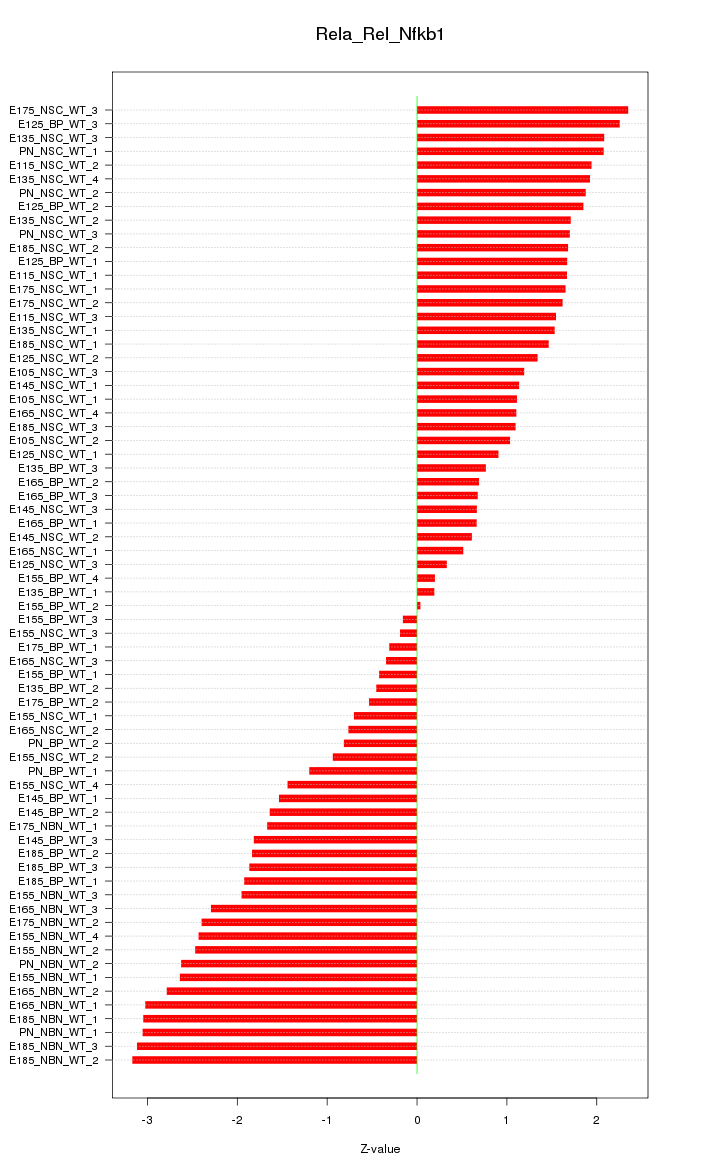
Motif ID: Rela_Rel_Nfkb1
Z-value: 1.683
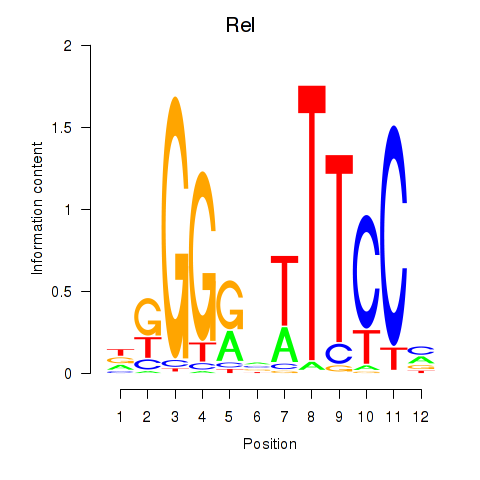
Transcription factors associated with Rela_Rel_Nfkb1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Nfkb1 | ENSMUSG00000028163.11 | Nfkb1 |
| Rel | ENSMUSG00000020275.8 | Rel |
| Rela | ENSMUSG00000024927.7 | Rela |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfkb1 | mm10_v2_chr3_-_135608221_135608290 | 0.82 | 1.9e-18 | Click! |
| Rela | mm10_v2_chr19_+_5637475_5637486 | 0.38 | 1.2e-03 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.8 | 47.5 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 4.6 | 27.5 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 3.8 | 19.0 | GO:0015671 | oxygen transport(GO:0015671) |
| 3.1 | 9.3 | GO:0072554 | arterial endothelial cell fate commitment(GO:0060844) blood vessel endothelial cell fate commitment(GO:0060846) endothelial cell fate specification(GO:0060847) Notch signaling pathway involved in arterial endothelial cell fate commitment(GO:0060853) blood vessel lumenization(GO:0072554) blood vessel endothelial cell fate specification(GO:0097101) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) positive regulation of heart induction(GO:1901321) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 2.7 | 13.7 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 2.5 | 10.2 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 2.5 | 7.6 | GO:0033860 | regulation of NAD(P)H oxidase activity(GO:0033860) |
| 2.5 | 7.5 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 2.4 | 9.6 | GO:1903416 | negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) regulation of calcidiol 1-monooxygenase activity(GO:0060558) response to glycoside(GO:1903416) |
| 2.4 | 7.1 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) negative regulation of interleukin-6-mediated signaling pathway(GO:0070104) |
| 2.1 | 21.3 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 2.1 | 14.4 | GO:0000022 | mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 2.0 | 20.4 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 2.0 | 21.8 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 2.0 | 11.9 | GO:0070094 | positive regulation of glucagon secretion(GO:0070094) |
| 1.8 | 9.2 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 1.8 | 7.0 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 1.8 | 5.3 | GO:0016115 | terpenoid catabolic process(GO:0016115) |
| 1.7 | 10.4 | GO:0002553 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 1.7 | 5.1 | GO:0015938 | coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) |
| 1.7 | 5.0 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 1.7 | 5.0 | GO:0090403 | regulation of purine nucleotide catabolic process(GO:0033121) negative regulation of purine nucleotide catabolic process(GO:0033122) regulation of fermentation(GO:0043465) oxidative stress-induced premature senescence(GO:0090403) negative regulation of fermentation(GO:1901003) |
| 1.7 | 5.0 | GO:0006116 | NADH oxidation(GO:0006116) |
| 1.7 | 6.6 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 1.5 | 6.1 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 1.5 | 4.5 | GO:0032916 | positive regulation of transforming growth factor beta3 production(GO:0032916) |
| 1.5 | 20.4 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 1.4 | 22.0 | GO:0002467 | germinal center formation(GO:0002467) |
| 1.3 | 24.7 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 1.2 | 8.6 | GO:0007296 | vitellogenesis(GO:0007296) |
| 1.2 | 5.9 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 1.1 | 3.4 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 1.1 | 13.3 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 1.1 | 5.5 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 1.1 | 8.7 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 1.0 | 3.1 | GO:0003349 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 1.0 | 4.1 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 1.0 | 6.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 1.0 | 3.0 | GO:0008228 | opsonization(GO:0008228) modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 1.0 | 9.0 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 1.0 | 2.9 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 1.0 | 9.8 | GO:1904707 | positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 1.0 | 4.8 | GO:2000157 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.9 | 0.9 | GO:0002678 | positive regulation of chronic inflammatory response(GO:0002678) |
| 0.9 | 11.2 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.9 | 2.8 | GO:0035622 | intrahepatic bile duct development(GO:0035622) cholangiocyte proliferation(GO:1990705) |
| 0.9 | 4.6 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.9 | 2.6 | GO:0030321 | transepithelial chloride transport(GO:0030321) transepithelial ammonium transport(GO:0070634) |
| 0.7 | 3.0 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.7 | 8.0 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.7 | 7.9 | GO:2000254 | regulation of male germ cell proliferation(GO:2000254) |
| 0.7 | 4.3 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.7 | 3.5 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.7 | 3.5 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.7 | 4.2 | GO:1902373 | negative regulation of mRNA catabolic process(GO:1902373) |
| 0.7 | 0.7 | GO:0010896 | regulation of triglyceride catabolic process(GO:0010896) positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.7 | 6.0 | GO:0042095 | interferon-gamma biosynthetic process(GO:0042095) |
| 0.7 | 4.0 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.7 | 3.3 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.7 | 2.0 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.6 | 8.4 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.6 | 1.9 | GO:0098501 | polynucleotide dephosphorylation(GO:0098501) |
| 0.6 | 5.0 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.6 | 3.1 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.6 | 1.8 | GO:1900275 | negative regulation of phospholipase C activity(GO:1900275) |
| 0.6 | 4.9 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.6 | 1.8 | GO:0040019 | positive regulation of embryonic development(GO:0040019) |
| 0.6 | 0.6 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.6 | 6.0 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.6 | 1.8 | GO:0060023 | soft palate development(GO:0060023) |
| 0.6 | 1.7 | GO:0060536 | cartilage morphogenesis(GO:0060536) |
| 0.6 | 2.3 | GO:0032382 | positive regulation of intracellular lipid transport(GO:0032379) positive regulation of intracellular sterol transport(GO:0032382) positive regulation of intracellular cholesterol transport(GO:0032385) lipid hydroperoxide transport(GO:1901373) positive regulation of cholesterol import(GO:1904109) positive regulation of sterol import(GO:2000911) |
| 0.6 | 9.6 | GO:1900153 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.5 | 2.2 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.5 | 9.8 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.5 | 2.7 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.5 | 9.0 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.5 | 1.1 | GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 0.5 | 2.1 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.5 | 3.6 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.5 | 7.6 | GO:0050434 | positive regulation of viral transcription(GO:0050434) |
| 0.5 | 5.1 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.5 | 1.5 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.5 | 4.5 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.5 | 0.5 | GO:1902460 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.5 | 8.5 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.5 | 3.3 | GO:0051004 | regulation of lipoprotein lipase activity(GO:0051004) |
| 0.5 | 12.2 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.5 | 1.4 | GO:2000852 | regulation of corticosterone secretion(GO:2000852) |
| 0.4 | 2.7 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.4 | 2.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.4 | 4.4 | GO:1901409 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.4 | 6.9 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.4 | 3.4 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.4 | 2.1 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.4 | 2.9 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.4 | 0.8 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.4 | 2.5 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.4 | 1.6 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.4 | 1.6 | GO:0090209 | mammary gland branching involved in pregnancy(GO:0060745) negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.4 | 4.9 | GO:0050965 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.4 | 1.9 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.4 | 7.8 | GO:0033622 | integrin activation(GO:0033622) |
| 0.4 | 1.5 | GO:0035553 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.4 | 2.9 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.3 | 0.7 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.3 | 2.4 | GO:0034433 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.3 | 1.0 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.3 | 2.9 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.3 | 1.3 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.3 | 18.5 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.3 | 2.8 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.3 | 2.7 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.3 | 1.8 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.3 | 4.2 | GO:0042182 | ketone catabolic process(GO:0042182) |
| 0.3 | 2.7 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.3 | 5.8 | GO:0030855 | epithelial cell differentiation(GO:0030855) |
| 0.3 | 14.5 | GO:0002066 | columnar/cuboidal epithelial cell development(GO:0002066) |
| 0.3 | 2.6 | GO:0043173 | nucleotide salvage(GO:0043173) |
| 0.3 | 2.0 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.3 | 1.9 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.3 | 3.3 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.3 | 0.8 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 0.3 | 1.6 | GO:1902998 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of tau-protein kinase activity(GO:1902949) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.3 | 4.7 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.3 | 1.8 | GO:2000370 | positive regulation of clathrin-mediated endocytosis(GO:2000370) |
| 0.3 | 1.3 | GO:0033136 | serine phosphorylation of STAT3 protein(GO:0033136) |
| 0.3 | 5.8 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.2 | 1.7 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.2 | 1.0 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.2 | 9.1 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.2 | 2.2 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.2 | 4.9 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.2 | 1.8 | GO:0009181 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.2 | 1.1 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.2 | 0.9 | GO:0070633 | transepithelial transport(GO:0070633) |
| 0.2 | 2.4 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.2 | 1.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.2 | 2.6 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.2 | 0.2 | GO:0060664 | epithelial cell proliferation involved in salivary gland morphogenesis(GO:0060664) |
| 0.2 | 2.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.2 | 1.7 | GO:1904816 | positive regulation of protein localization to chromosome, telomeric region(GO:1904816) |
| 0.2 | 1.9 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.2 | 1.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.2 | 1.6 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.2 | 2.2 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.2 | 0.6 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.2 | 0.8 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.2 | 1.6 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.2 | 2.9 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.2 | 1.7 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.2 | 0.4 | GO:0097527 | necroptotic signaling pathway(GO:0097527) |
| 0.2 | 1.8 | GO:0071435 | stabilization of membrane potential(GO:0030322) potassium ion export(GO:0071435) |
| 0.2 | 1.2 | GO:1903867 | chorion development(GO:0060717) extraembryonic membrane development(GO:1903867) |
| 0.2 | 0.5 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.2 | 6.0 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.2 | 6.5 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.2 | 1.2 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.2 | 0.2 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.2 | 0.8 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.2 | 1.1 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.2 | 0.5 | GO:1904884 | telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.2 | 2.1 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.2 | 0.3 | GO:1904749 | regulation of protein localization to nucleolus(GO:1904749) positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.2 | 0.9 | GO:1903912 | negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:1903912) |
| 0.2 | 5.3 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.1 | 0.6 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 1.6 | GO:0006337 | nucleosome disassembly(GO:0006337) protein-DNA complex disassembly(GO:0032986) |
| 0.1 | 0.9 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.1 | 6.3 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.1 | 3.4 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 2.7 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.1 | 1.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 0.9 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 0.4 | GO:1902528 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.1 | 0.8 | GO:0031953 | negative regulation of protein autophosphorylation(GO:0031953) |
| 0.1 | 0.5 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.1 | 0.6 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.1 | 0.4 | GO:0033030 | negative regulation of neutrophil apoptotic process(GO:0033030) |
| 0.1 | 2.9 | GO:0035115 | embryonic forelimb morphogenesis(GO:0035115) |
| 0.1 | 0.2 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.7 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.1 | 2.1 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.1 | 1.7 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 4.6 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.1 | 5.8 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.1 | 6.7 | GO:0071774 | response to fibroblast growth factor(GO:0071774) |
| 0.1 | 3.4 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.1 | 1.7 | GO:0045672 | positive regulation of osteoclast differentiation(GO:0045672) |
| 0.1 | 0.4 | GO:0031509 | telomeric heterochromatin assembly(GO:0031509) regulation of chromosome condensation(GO:0060623) negative regulation of chromosome condensation(GO:1902340) |
| 0.1 | 0.4 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.1 | 0.4 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.1 | 0.4 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.1 | 1.6 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.1 | 1.0 | GO:0044380 | protein localization to cytoskeleton(GO:0044380) |
| 0.1 | 0.3 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.1 | 1.3 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.1 | 0.3 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.1 | 15.9 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.1 | 2.6 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.3 | GO:0034035 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) |
| 0.1 | 0.8 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.1 | 2.7 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.1 | 4.4 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.1 | 0.4 | GO:0030836 | positive regulation of actin filament depolymerization(GO:0030836) |
| 0.1 | 2.2 | GO:0042130 | negative regulation of T cell proliferation(GO:0042130) |
| 0.1 | 1.8 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.1 | 3.2 | GO:0048661 | positive regulation of smooth muscle cell proliferation(GO:0048661) |
| 0.1 | 3.0 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.1 | 0.6 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 1.6 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 2.3 | GO:0031365 | N-terminal protein amino acid modification(GO:0031365) |
| 0.1 | 2.7 | GO:0001501 | skeletal system development(GO:0001501) |
| 0.1 | 2.4 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 0.7 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.1 | 3.6 | GO:0060021 | palate development(GO:0060021) |
| 0.1 | 0.3 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.1 | 1.5 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 0.3 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.1 | 1.6 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.1 | 1.1 | GO:0033233 | regulation of protein sumoylation(GO:0033233) positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.8 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 1.4 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 0.7 | GO:1902287 | semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 1.3 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.0 | 3.0 | GO:2001237 | negative regulation of extrinsic apoptotic signaling pathway(GO:2001237) |
| 0.0 | 0.9 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 1.1 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 1.1 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.0 | 0.2 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.3 | GO:0006532 | aspartate biosynthetic process(GO:0006532) |
| 0.0 | 0.5 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.0 | 1.9 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.6 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 1.1 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.4 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 2.2 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.9 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 1.2 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.4 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.1 | GO:0048146 | positive regulation of fibroblast proliferation(GO:0048146) |
| 0.0 | 0.1 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 1.1 | GO:0002209 | behavioral fear response(GO:0001662) behavioral defense response(GO:0002209) |
| 0.0 | 0.0 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.0 | 0.1 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.0 | 0.3 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 1.5 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.4 | GO:0042035 | regulation of cytokine biosynthetic process(GO:0042035) |
| 0.0 | 0.6 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.2 | GO:1904407 | positive regulation of nitric oxide biosynthetic process(GO:0045429) positive regulation of nitric oxide metabolic process(GO:1904407) |
| 0.0 | 0.0 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 1.5 | GO:0001822 | kidney development(GO:0001822) |
| 0.0 | 0.1 | GO:0031274 | regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.2 | GO:0060441 | epithelial tube branching involved in lung morphogenesis(GO:0060441) |
| 0.0 | 0.0 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.4 | 37.8 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 2.9 | 23.5 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 2.4 | 19.0 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 2.2 | 8.7 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 2.1 | 6.3 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 1.9 | 9.3 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 1.8 | 10.6 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 1.7 | 8.7 | GO:0031523 | Myb complex(GO:0031523) |
| 1.7 | 13.7 | GO:0098536 | deuterosome(GO:0098536) |
| 1.5 | 6.0 | GO:0043293 | apoptosome(GO:0043293) |
| 1.4 | 4.3 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 1.4 | 5.7 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 1.4 | 5.5 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 1.3 | 27.5 | GO:0043657 | host(GO:0018995) host cell part(GO:0033643) host cell(GO:0043657) |
| 1.1 | 16.4 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 1.0 | 2.0 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 1.0 | 3.9 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) insulin-like growth factor ternary complex(GO:0042567) |
| 1.0 | 10.5 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.9 | 23.7 | GO:0002102 | podosome(GO:0002102) |
| 0.9 | 4.5 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.9 | 2.6 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.8 | 6.6 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.8 | 5.3 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.7 | 2.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.6 | 3.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.6 | 1.7 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.6 | 5.0 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.5 | 1.6 | GO:0030690 | Noc1p-Noc2p complex(GO:0030690) |
| 0.5 | 17.7 | GO:0043034 | costamere(GO:0043034) |
| 0.5 | 1.4 | GO:0005595 | collagen type XII trimer(GO:0005595) anchoring collagen complex(GO:0030934) |
| 0.5 | 3.7 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.4 | 5.6 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.4 | 2.6 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.3 | 2.0 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.3 | 4.2 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.3 | 4.8 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.3 | 1.6 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.3 | 1.7 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.3 | 6.5 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.3 | 1.9 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.3 | 6.0 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.3 | 4.4 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.3 | 11.5 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.3 | 4.2 | GO:0010369 | chromocenter(GO:0010369) |
| 0.3 | 6.9 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.3 | 31.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.2 | 17.9 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.2 | 1.6 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.2 | 1.6 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) neurofibrillary tangle(GO:0097418) |
| 0.2 | 1.1 | GO:0008623 | CHRAC(GO:0008623) |
| 0.2 | 11.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.2 | 1.8 | GO:0030478 | actin cap(GO:0030478) |
| 0.2 | 0.7 | GO:0002944 | cyclin K-CDK12 complex(GO:0002944) |
| 0.2 | 4.1 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.2 | 4.6 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.2 | 4.1 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.2 | 1.1 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.2 | 0.7 | GO:0016012 | dystroglycan complex(GO:0016011) sarcoglycan complex(GO:0016012) |
| 0.2 | 1.7 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.2 | 2.0 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.2 | 1.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.2 | 3.4 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.2 | 1.5 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.2 | 7.0 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.2 | 0.5 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.2 | 5.0 | GO:0005903 | brush border(GO:0005903) |
| 0.2 | 1.0 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.2 | 1.3 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.2 | 1.7 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.2 | 2.0 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.2 | 0.9 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.1 | 17.0 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 4.7 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.1 | 1.8 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 0.5 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 1.4 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 0.7 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 1.0 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 1.6 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 7.7 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.1 | 7.7 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 1.4 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 1.8 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 0.4 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.1 | 4.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 2.5 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 1.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 3.8 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 7.4 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 4.7 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 8.6 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.1 | 0.4 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.1 | 3.5 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 6.2 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 6.3 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 3.8 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 0.9 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.4 | GO:0001740 | Barr body(GO:0001740) |
| 0.1 | 1.0 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.1 | 0.2 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 1.0 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.1 | 1.4 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 11.4 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.1 | 0.7 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 12.1 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 1.8 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 24.4 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.1 | 1.9 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 0.4 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 0.9 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 1.0 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 1.0 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 0.7 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 1.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 0.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.4 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 1.5 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 1.6 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 14.5 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 3.1 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 2.5 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 5.7 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 1.2 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 0.1 | GO:0044302 | dentate gyrus mossy fiber(GO:0044302) |
| 0.0 | 0.6 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 1.1 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 2.5 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 2.4 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 1.6 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.4 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 1.0 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 4.2 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 1.1 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.4 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 2.0 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.1 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 1.1 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.7 | 19.0 | GO:0031721 | hemoglobin alpha binding(GO:0031721) |
| 4.3 | 12.9 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 2.7 | 27.5 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 2.5 | 7.5 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 2.2 | 8.7 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 2.2 | 8.7 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 2.0 | 6.1 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) oncostatin-M receptor activity(GO:0004924) |
| 1.7 | 20.4 | GO:0031996 | thioesterase binding(GO:0031996) |
| 1.7 | 5.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 1.6 | 6.5 | GO:1990254 | keratin filament binding(GO:1990254) |
| 1.5 | 13.3 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 1.4 | 4.2 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 1.3 | 5.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 1.1 | 5.7 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 1.1 | 3.4 | GO:0016880 | acid-ammonia (or amide) ligase activity(GO:0016880) |
| 1.1 | 4.5 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 1.0 | 3.1 | GO:0004368 | glycerol-3-phosphate dehydrogenase activity(GO:0004368) |
| 0.9 | 3.6 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.8 | 9.2 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.8 | 3.3 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.8 | 4.8 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.7 | 5.1 | GO:0070191 | methionine-R-sulfoxide reductase activity(GO:0070191) |
| 0.7 | 3.6 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.7 | 12.0 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.7 | 6.9 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.7 | 7.6 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.6 | 9.0 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.6 | 1.2 | GO:0046978 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.6 | 11.5 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.6 | 6.6 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.6 | 1.8 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.6 | 6.0 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.6 | 4.1 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.6 | 2.3 | GO:0033814 | propanoyl-CoA C-acyltransferase activity(GO:0033814) propionyl-CoA C2-trimethyltridecanoyltransferase activity(GO:0050632) phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.6 | 2.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.6 | 3.9 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.6 | 4.5 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.6 | 22.3 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.5 | 4.9 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.5 | 2.7 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.5 | 1.6 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.5 | 3.5 | GO:0015288 | porin activity(GO:0015288) |
| 0.5 | 4.5 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.5 | 3.0 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.5 | 5.3 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.5 | 2.4 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.5 | 1.9 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) |
| 0.5 | 13.0 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.5 | 5.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.5 | 1.4 | GO:0031779 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.4 | 1.8 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.4 | 4.3 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.4 | 1.3 | GO:0008732 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.4 | 33.3 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.4 | 1.6 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.4 | 5.6 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.4 | 1.9 | GO:0046403 | polynucleotide 3'-phosphatase activity(GO:0046403) |
| 0.4 | 4.9 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.4 | 1.5 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.4 | 2.6 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.4 | 10.3 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.3 | 5.6 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.3 | 4.6 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.3 | 1.6 | GO:0051880 | Y-form DNA binding(GO:0000403) bubble DNA binding(GO:0000405) G-quadruplex DNA binding(GO:0051880) |
| 0.3 | 2.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.3 | 2.2 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.3 | 3.7 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.3 | 1.5 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.3 | 0.9 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.3 | 2.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.3 | 30.9 | GO:0005178 | integrin binding(GO:0005178) |
| 0.3 | 0.8 | GO:0034437 | glycoprotein transporter activity(GO:0034437) |
| 0.3 | 1.6 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.3 | 2.6 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.2 | 2.7 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.2 | 2.7 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.2 | 14.9 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.2 | 1.8 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.2 | 1.4 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.2 | 6.0 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.2 | 33.6 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.2 | 1.6 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.2 | 1.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.2 | 0.6 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.2 | 11.2 | GO:0019003 | GDP binding(GO:0019003) |
| 0.2 | 0.5 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.2 | 7.2 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.2 | 2.9 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 7.4 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.2 | 1.2 | GO:0045703 | pinocarveol dehydrogenase activity(GO:0018446) chloral hydrate dehydrogenase activity(GO:0018447) hydroxymethylmethylsilanediol oxidase activity(GO:0018448) 1-phenylethanol dehydrogenase activity(GO:0018449) myrtenol dehydrogenase activity(GO:0018450) cis-1,2-dihydroxy-1,2-dihydro-8-carboxynaphthalene dehydrogenase activity(GO:0034522) 3-hydroxy-4-methyloctanoyl-CoA dehydrogenase activity(GO:0034582) 2-hydroxy-4-isopropenylcyclohexane-1-carboxyl-CoA dehydrogenase activity(GO:0034778) cis-9,10-dihydroanthracene-9,10-diol dehydrogenase activity(GO:0034817) citronellol dehydrogenase activity(GO:0034821) naphthyl-2-hydroxymethyl-succinyl-CoA dehydrogenase activity(GO:0034847) 2,4,4-trimethyl-1-pentanol dehydrogenase activity(GO:0034863) 2,4,4-trimethyl-3-hydroxypentanoyl-CoA dehydrogenase activity(GO:0034868) 1-hydroxy-4,4-dimethylpentan-3-one dehydrogenase activity(GO:0034871) endosulfan diol dehydrogenase activity(GO:0034891) endosulfan hydroxyether dehydrogenase activity(GO:0034901) 3-hydroxy-2-methylhexanoyl-CoA dehydrogenase activity(GO:0034918) 3-hydroxy-2,6-dimethyl-5-methylene-heptanoyl-CoA dehydrogenase activity(GO:0034944) versicolorin reductase activity(GO:0042469) ketoreductase activity(GO:0045703) |
| 0.2 | 0.5 | GO:0032405 | MutLalpha complex binding(GO:0032405) |
| 0.2 | 0.9 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.2 | 7.7 | GO:0020037 | heme binding(GO:0020037) |
| 0.1 | 2.2 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 1.0 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 8.4 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.1 | 3.7 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 1.8 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 72.6 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) |
| 0.1 | 0.4 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) C-X-C chemokine binding(GO:0019958) |
| 0.1 | 2.1 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 1.0 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 0.4 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 3.6 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 5.8 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.1 | 0.6 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 4.2 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.1 | 4.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.6 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 3.2 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.1 | 2.0 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 0.3 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.1 | 0.7 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 6.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 0.3 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) |
| 0.1 | 4.7 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 1.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 2.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 1.9 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 0.9 | GO:0034951 | pivalyl-CoA mutase activity(GO:0034784) o-hydroxylaminobenzoate mutase activity(GO:0034951) lupeol synthase activity(GO:0042299) beta-amyrin synthase activity(GO:0042300) baruol synthase activity(GO:0080011) |
| 0.1 | 1.1 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 2.8 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 1.9 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 0.4 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.1 | 1.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.6 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 3.0 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 1.2 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.1 | 1.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 27.6 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 1.1 | GO:0050253 | prenylcysteine methylesterase activity(GO:0010296) 1-oxa-2-oxocycloheptane lactonase activity(GO:0018731) sulfolactone hydrolase activity(GO:0018732) 3,4-dihydrocoumarin hydrolase activity(GO:0018733) butyrolactone hydrolase activity(GO:0018734) endosulfan lactone lactonase activity(GO:0034892) L-ascorbate 6-phosphate lactonase activity(GO:0035460) Ser-tRNA(Thr) hydrolase activity(GO:0043905) Ala-tRNA(Pro) hydrolase activity(GO:0043906) Cys-tRNA(Pro) hydrolase activity(GO:0043907) Ser(Gly)-tRNA(Ala) hydrolase activity(GO:0043908) all-trans-retinyl-palmitate hydrolase, all-trans-retinol forming activity(GO:0047376) retinyl-palmitate esterase activity(GO:0050253) mannosyl-oligosaccharide 1,6-alpha-mannosidase activity(GO:0052767) mannosyl-oligosaccharide 1,3-alpha-mannosidase activity(GO:0052768) methyl indole-3-acetate esterase activity(GO:0080030) methyl salicylate esterase activity(GO:0080031) methyl jasmonate esterase activity(GO:0080032) |
| 0.1 | 0.6 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.1 | 0.2 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.1 | 2.4 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 0.2 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.1 | 2.2 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.1 | 0.9 | GO:0030553 | cGMP binding(GO:0030553) 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 0.7 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 1.0 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 1.1 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 0.7 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.5 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 1.7 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 0.8 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 1.7 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.7 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 9.2 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.3 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) poly(G) binding(GO:0034046) histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 13.5 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.3 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.0 | 4.5 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 0.0 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 1.4 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 1.3 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 1.7 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.5 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.4 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.4 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.2 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.4 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.6 | GO:0017171 | serine-type peptidase activity(GO:0008236) serine hydrolase activity(GO:0017171) |
| 0.0 | 0.4 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 1.0 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 1.2 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 20.7 | GO:0044822 | poly(A) RNA binding(GO:0044822) |
| 0.0 | 1.3 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 1.1 | GO:0003735 | structural constituent of ribosome(GO:0003735) |