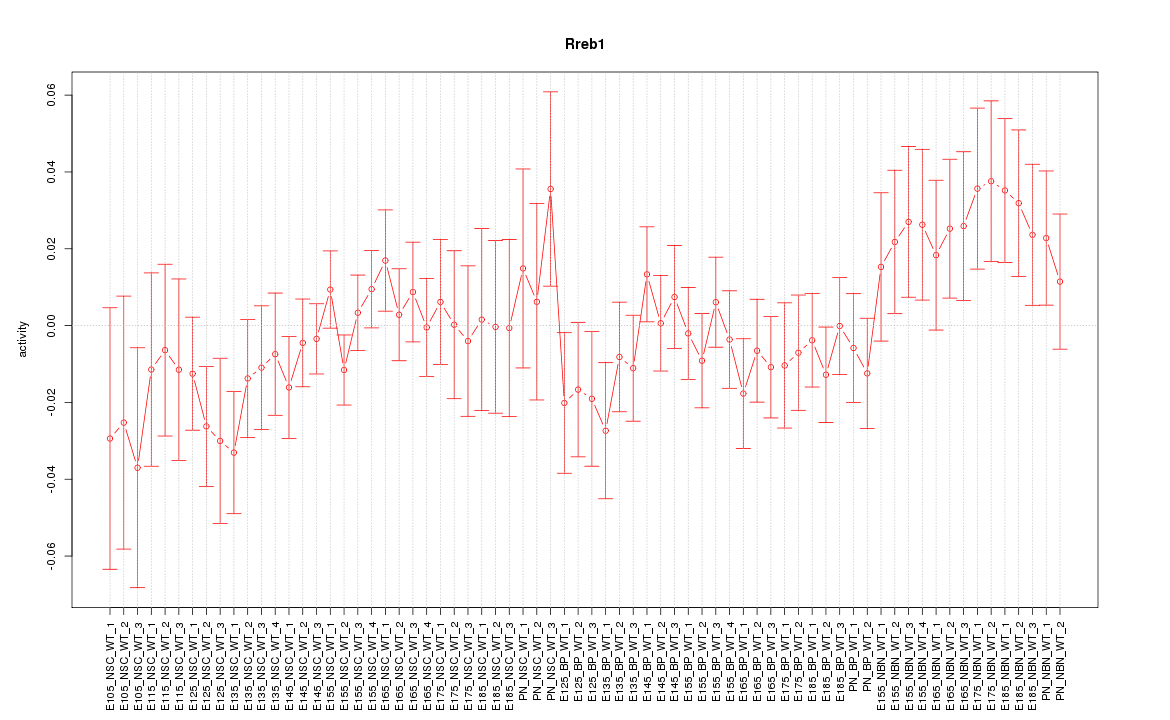
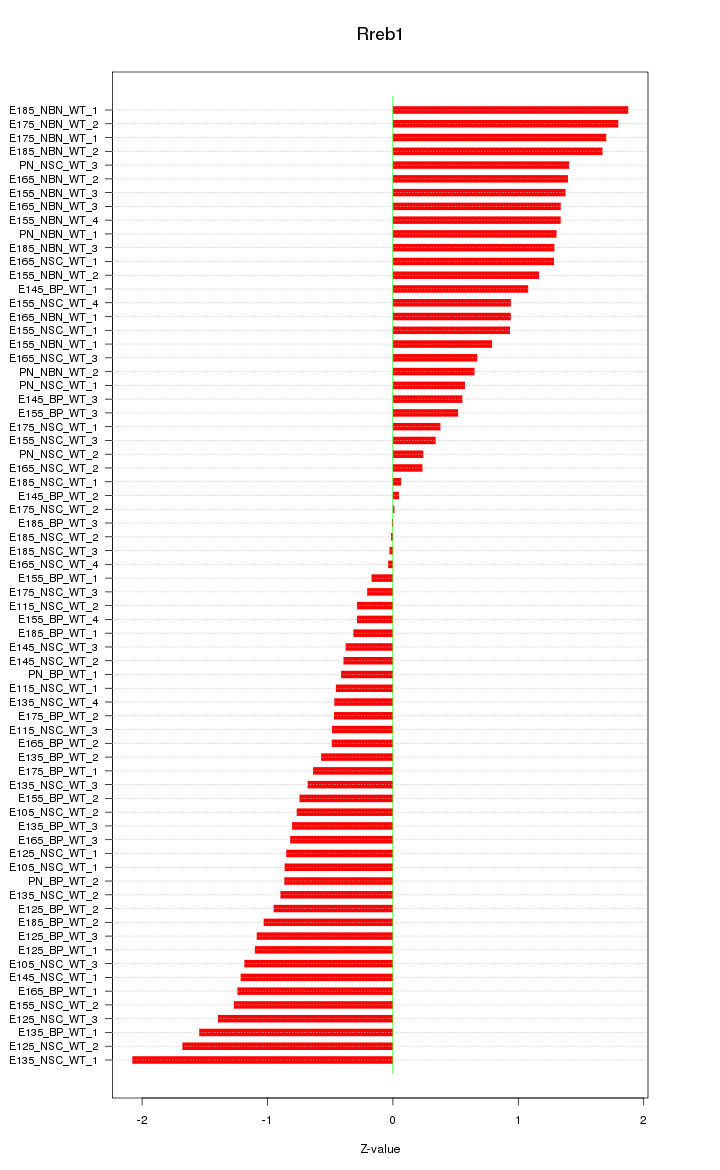
Motif ID: Rreb1
Z-value: 0.966
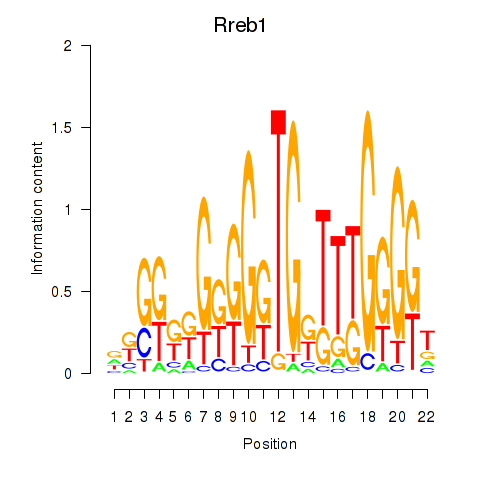
Transcription factors associated with Rreb1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Rreb1 | ENSMUSG00000039087.10 | Rreb1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rreb1 | mm10_v2_chr13_+_37826225_37826315 | -0.09 | 4.5e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 10.7 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 2.0 | 6.0 | GO:0030827 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 1.9 | 9.3 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 1.4 | 4.2 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 1.3 | 4.0 | GO:1904049 | regulation of spontaneous neurotransmitter secretion(GO:1904048) negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 1.3 | 7.8 | GO:1903056 | regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 1.2 | 5.8 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 1.1 | 5.7 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 1.1 | 4.3 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.9 | 4.3 | GO:1900085 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) |
| 0.8 | 4.0 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.8 | 2.4 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.8 | 2.4 | GO:1902528 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.7 | 2.1 | GO:0048686 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.7 | 5.6 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.7 | 4.1 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.7 | 2.7 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.7 | 2.0 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.6 | 3.4 | GO:0019336 | phenol-containing compound catabolic process(GO:0019336) |
| 0.6 | 11.0 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.5 | 1.6 | GO:0042939 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.5 | 2.7 | GO:0034141 | positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.5 | 7.7 | GO:0043045 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.5 | 1.9 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.5 | 0.5 | GO:0034145 | positive regulation of toll-like receptor 4 signaling pathway(GO:0034145) |
| 0.4 | 2.7 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.4 | 3.9 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.4 | 3.0 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.4 | 1.7 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.4 | 0.4 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.4 | 2.1 | GO:1902856 | negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 0.4 | 1.7 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.4 | 2.4 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.4 | 1.6 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) Harderian gland development(GO:0070384) |
| 0.4 | 1.6 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.4 | 1.6 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.4 | 1.2 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.3 | 3.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.3 | 1.0 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) |
| 0.3 | 1.9 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 0.3 | 1.0 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.3 | 1.9 | GO:0061718 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glycolytic process through glucose-1-phosphate(GO:0061622) glucose catabolic process to pyruvate(GO:0061718) |
| 0.3 | 1.2 | GO:1990035 | calcium ion import into cell(GO:1990035) |
| 0.3 | 0.8 | GO:0046077 | dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) deoxyribonucleoside diphosphate biosynthetic process(GO:0009189) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 0.3 | 1.1 | GO:1900110 | negative regulation of histone H3-K9 dimethylation(GO:1900110) |
| 0.3 | 7.6 | GO:0022401 | desensitization of G-protein coupled receptor protein signaling pathway(GO:0002029) negative adaptation of signaling pathway(GO:0022401) |
| 0.2 | 1.2 | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.2 | 1.9 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.2 | 1.6 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.2 | 0.9 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.2 | 9.9 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.2 | 1.6 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.2 | 3.1 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.2 | 0.9 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.2 | 0.7 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.2 | 3.5 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.2 | 1.7 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.2 | 0.6 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.2 | 1.3 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.2 | 1.0 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.2 | 0.8 | GO:1900242 | regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.2 | 1.5 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.2 | 6.6 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.2 | 1.6 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.2 | 2.3 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.2 | 0.8 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.2 | 2.7 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.2 | 0.7 | GO:0044330 | canonical Wnt signaling pathway involved in positive regulation of wound healing(GO:0044330) lactic acid secretion(GO:0046722) regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.2 | 0.5 | GO:0002554 | serotonin secretion by platelet(GO:0002554) interleukin-3 production(GO:0032632) beta selection(GO:0043366) |
| 0.2 | 0.7 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.2 | 1.5 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.2 | 1.7 | GO:0021955 | central nervous system neuron axonogenesis(GO:0021955) |
| 0.2 | 0.2 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.2 | 1.8 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.2 | 1.3 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.2 | 0.8 | GO:0090164 | asymmetric Golgi ribbon formation(GO:0090164) |
| 0.2 | 0.5 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.1 | 6.0 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.1 | 0.9 | GO:1900112 | regulation of histone H3-K9 trimethylation(GO:1900112) negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.1 | 6.4 | GO:0090004 | positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.1 | 2.9 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.1 | 1.5 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.1 | 0.7 | GO:2000987 | positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.1 | 2.9 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.1 | 1.2 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.1 | 1.2 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 0.8 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.1 | 1.1 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.1 | 2.1 | GO:0017144 | drug metabolic process(GO:0017144) |
| 0.1 | 0.6 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.1 | 2.6 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.1 | 0.9 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 3.5 | GO:2000463 | positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.1 | 0.3 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.1 | 18.7 | GO:0051017 | actin filament bundle assembly(GO:0051017) |
| 0.1 | 1.2 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.1 | 1.0 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.1 | 0.5 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.1 | 0.5 | GO:0048597 | post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.1 | 0.4 | GO:0010726 | positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.1 | 0.3 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.1 | 0.9 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 3.2 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.1 | 2.7 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 0.9 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 1.4 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 2.0 | GO:0051926 | negative regulation of calcium ion transport(GO:0051926) |
| 0.1 | 0.3 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.1 | 2.7 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.1 | 0.2 | GO:0046618 | drug export(GO:0046618) |
| 0.1 | 0.9 | GO:1901032 | negative regulation of response to reactive oxygen species(GO:1901032) negative regulation of hydrogen peroxide-induced cell death(GO:1903206) |
| 0.1 | 0.3 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.1 | 0.2 | GO:0051342 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.1 | 0.9 | GO:0097484 | dendrite extension(GO:0097484) |
| 0.1 | 9.7 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.1 | 0.9 | GO:0032469 | endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.1 | 0.8 | GO:0072010 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.1 | 0.8 | GO:0003283 | atrial septum development(GO:0003283) |
| 0.1 | 0.2 | GO:0001865 | NK T cell differentiation(GO:0001865) antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) |
| 0.1 | 0.5 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.1 | 0.1 | GO:0070346 | positive regulation of fat cell proliferation(GO:0070346) |
| 0.1 | 2.4 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.1 | 1.0 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 7.1 | GO:0071805 | potassium ion transmembrane transport(GO:0071805) |
| 0.1 | 0.3 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.4 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.4 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.0 | 0.4 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 1.0 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 2.1 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.0 | 2.5 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.1 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.0 | 1.3 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.9 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 1.3 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.0 | 0.4 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.2 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.3 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.0 | 1.5 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.5 | GO:0008209 | androgen metabolic process(GO:0008209) |
| 0.0 | 0.6 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.4 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 3.9 | GO:0007051 | spindle organization(GO:0007051) |
| 0.0 | 0.3 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.0 | 0.1 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.4 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.8 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 1.5 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.4 | GO:0042775 | mitochondrial ATP synthesis coupled electron transport(GO:0042775) |
| 0.0 | 0.0 | GO:0030826 | regulation of cGMP biosynthetic process(GO:0030826) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.8 | GO:0006813 | potassium ion transport(GO:0006813) |
| 0.0 | 0.2 | GO:0044406 | adhesion of symbiont to host(GO:0044406) |
| 0.0 | 0.7 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.4 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.0 | GO:0070305 | response to cGMP(GO:0070305) cellular response to cGMP(GO:0071321) |
| 0.0 | 0.6 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.6 | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.0 | 2.0 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.0 | 0.2 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.0 | 1.0 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 1.6 | GO:0006633 | fatty acid biosynthetic process(GO:0006633) |
| 0.0 | 0.4 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 1.6 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 0.5 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 1.7 | GO:0051321 | meiotic cell cycle(GO:0051321) |
| 0.0 | 0.3 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) positive regulation of response to extracellular stimulus(GO:0032106) positive regulation of response to nutrient levels(GO:0032109) |
| 0.0 | 6.9 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 0.3 | GO:0007608 | sensory perception of smell(GO:0007608) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.2 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.8 | 4.0 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.6 | 6.0 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.5 | 13.7 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.5 | 1.4 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.5 | 1.9 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.4 | 5.7 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.4 | 2.2 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.4 | 1.9 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.4 | 5.8 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.3 | 2.7 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.3 | 4.3 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.3 | 9.6 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.3 | 1.8 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.3 | 4.1 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.2 | 4.0 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.2 | 1.4 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.2 | 0.7 | GO:0098984 | neuron to neuron synapse(GO:0098984) |
| 0.2 | 7.8 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.2 | 2.4 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.2 | 1.9 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.2 | 11.8 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.2 | 0.8 | GO:1990357 | terminal web(GO:1990357) |
| 0.2 | 1.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 0.9 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.1 | 1.0 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 2.4 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 1.5 | GO:0042599 | lamellar body(GO:0042599) |
| 0.1 | 0.4 | GO:1990879 | CST complex(GO:1990879) |
| 0.1 | 1.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 1.2 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 1.7 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 0.5 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 5.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 1.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.7 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.1 | 4.0 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 12.8 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 1.9 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 1.0 | GO:0001527 | microfibril(GO:0001527) |
| 0.1 | 0.5 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.1 | 2.1 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 3.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 1.5 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 1.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.0 | 1.0 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 6.2 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.0 | 2.9 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 8.6 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 0.8 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 1.3 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.4 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.9 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 6.0 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.0 | 4.3 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.4 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.1 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.9 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 1.3 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 3.8 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.4 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 1.0 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.2 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 4.2 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 1.3 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.8 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 1.2 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 0.9 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 1.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.2 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.0 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.5 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 3.3 | GO:0016604 | nuclear body(GO:0016604) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.6 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 1.9 | 5.7 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) glycoprotein transporter activity(GO:0034437) |
| 1.8 | 10.7 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 1.0 | 3.9 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 1.0 | 6.8 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.9 | 2.8 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.9 | 4.3 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.8 | 2.5 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.8 | 4.6 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.7 | 4.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.6 | 1.9 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.5 | 1.5 | GO:0051379 | epinephrine binding(GO:0051379) |
| 0.5 | 1.9 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.4 | 2.2 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.4 | 2.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.4 | 2.0 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.4 | 1.2 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.4 | 1.6 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.4 | 1.8 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.3 | 1.0 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.3 | 1.0 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 0.3 | 2.0 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.3 | 6.0 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.3 | 4.3 | GO:0070636 | single-strand selective uracil DNA N-glycosylase activity(GO:0017065) nicotinamide riboside hydrolase activity(GO:0070635) nicotinic acid riboside hydrolase activity(GO:0070636) deoxyribonucleoside 5'-monophosphate N-glycosidase activity(GO:0070694) |
| 0.3 | 6.5 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.3 | 9.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.3 | 1.9 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.3 | 1.3 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.3 | 4.6 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.3 | 1.7 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.3 | 0.8 | GO:0004127 | cytidylate kinase activity(GO:0004127) uridylate kinase activity(GO:0009041) |
| 0.3 | 4.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.3 | 3.1 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.3 | 8.8 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.2 | 1.6 | GO:0015651 | quaternary ammonium group transmembrane transporter activity(GO:0015651) |
| 0.2 | 1.1 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.2 | 4.2 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.2 | 5.1 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.2 | 2.0 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.2 | 0.7 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.2 | 1.6 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.2 | 0.6 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.2 | 0.9 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.2 | 1.2 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 1.6 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 2.4 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 1.4 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.1 | 1.2 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 1.7 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.1 | 1.7 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.1 | 7.5 | GO:0044824 | integrase activity(GO:0008907) T/G mismatch-specific endonuclease activity(GO:0043765) retroviral integrase activity(GO:0044823) retroviral 3' processing activity(GO:0044824) |
| 0.1 | 5.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 1.3 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 1.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.8 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.1 | 5.6 | GO:0004120 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) photoreceptor cyclic-nucleotide phosphodiesterase activity(GO:0004120) 7,8-dihydro-D-neopterin 2',3'-cyclic phosphate phosphodiesterase activity(GO:0044688) inositol phosphosphingolipid phospholipase activity(GO:0052712) inositol phosphorylceramide phospholipase activity(GO:0052713) mannosyl-inositol phosphorylceramide phospholipase activity(GO:0052714) mannosyl-diinositol phosphorylceramide phospholipase activity(GO:0052715) |
| 0.1 | 0.5 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.1 | 0.9 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 0.3 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.1 | 0.4 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.1 | 0.7 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 0.2 | GO:0036004 | GAF domain binding(GO:0036004) |
| 0.1 | 2.7 | GO:0034930 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) heparan sulfate 2-O-sulfotransferase activity(GO:0004394) HNK-1 sulfotransferase activity(GO:0016232) heparan sulfate 6-O-sulfotransferase activity(GO:0017095) trans-9R,10R-dihydrodiolphenanthrene sulfotransferase activity(GO:0018721) 1-phenanthrol sulfotransferase activity(GO:0018722) 3-phenanthrol sulfotransferase activity(GO:0018723) 4-phenanthrol sulfotransferase activity(GO:0018724) trans-3,4-dihydrodiolphenanthrene sulfotransferase activity(GO:0018725) 9-phenanthrol sulfotransferase activity(GO:0018726) 2-phenanthrol sulfotransferase activity(GO:0018727) phenanthrol sulfotransferase activity(GO:0019111) 1-hydroxypyrene sulfotransferase activity(GO:0034930) proteoglycan sulfotransferase activity(GO:0050698) cholesterol sulfotransferase activity(GO:0051922) hydroxyjasmonate sulfotransferase activity(GO:0080131) |
| 0.1 | 2.7 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 1.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 0.5 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 3.0 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 0.7 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.1 | 0.5 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.1 | 0.3 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.1 | 9.3 | GO:0008026 | ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.1 | 1.1 | GO:0001163 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.1 | 11.9 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 2.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 1.0 | GO:0023026 | MHC protein complex binding(GO:0023023) MHC class II protein complex binding(GO:0023026) |
| 0.1 | 1.3 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 2.4 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 1.5 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.1 | 0.8 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.1 | 0.4 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.1 | 0.4 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 1.6 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 1.9 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.1 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 1.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 2.4 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.9 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.3 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.5 | GO:0022842 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.2 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 6.9 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 1.5 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 1.7 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 4.2 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 1.7 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 3.7 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 1.9 | GO:0042562 | hormone binding(GO:0042562) |
| 0.0 | 2.6 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 5.9 | GO:0005088 | Ras guanyl-nucleotide exchange factor activity(GO:0005088) |
| 0.0 | 1.4 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.3 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 9.0 | GO:0043774 | UDP-N-acetylmuramoylalanyl-D-glutamyl-2,6-diaminopimelate-D-alanyl-D-alanine ligase activity(GO:0008766) ribosomal S6-glutamic acid ligase activity(GO:0018169) coenzyme F420-0 gamma-glutamyl ligase activity(GO:0043773) coenzyme F420-2 alpha-glutamyl ligase activity(GO:0043774) protein-glycine ligase activity(GO:0070735) protein-glycine ligase activity, initiating(GO:0070736) protein-glycine ligase activity, elongating(GO:0070737) tubulin-glycine ligase activity(GO:0070738) |
| 0.0 | 0.9 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 2.2 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.2 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.9 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 1.2 | GO:0061659 | ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.0 | 0.1 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 1.6 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.2 | GO:1901682 | sulfur compound transmembrane transporter activity(GO:1901682) |
| 0.0 | 1.1 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 2.9 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.4 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.6 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 2.7 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 0.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.6 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 3.7 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.1 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.1 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 3.5 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 0.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.6 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.2 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.1 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |