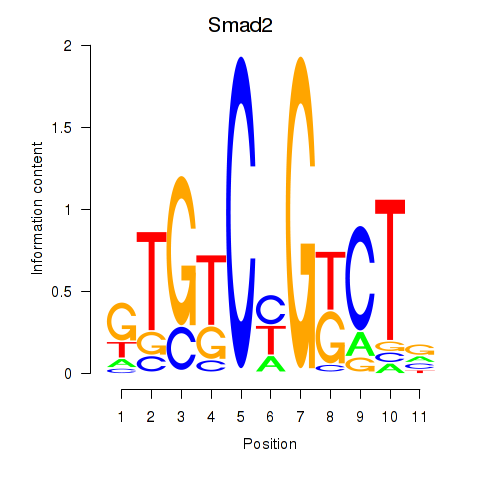
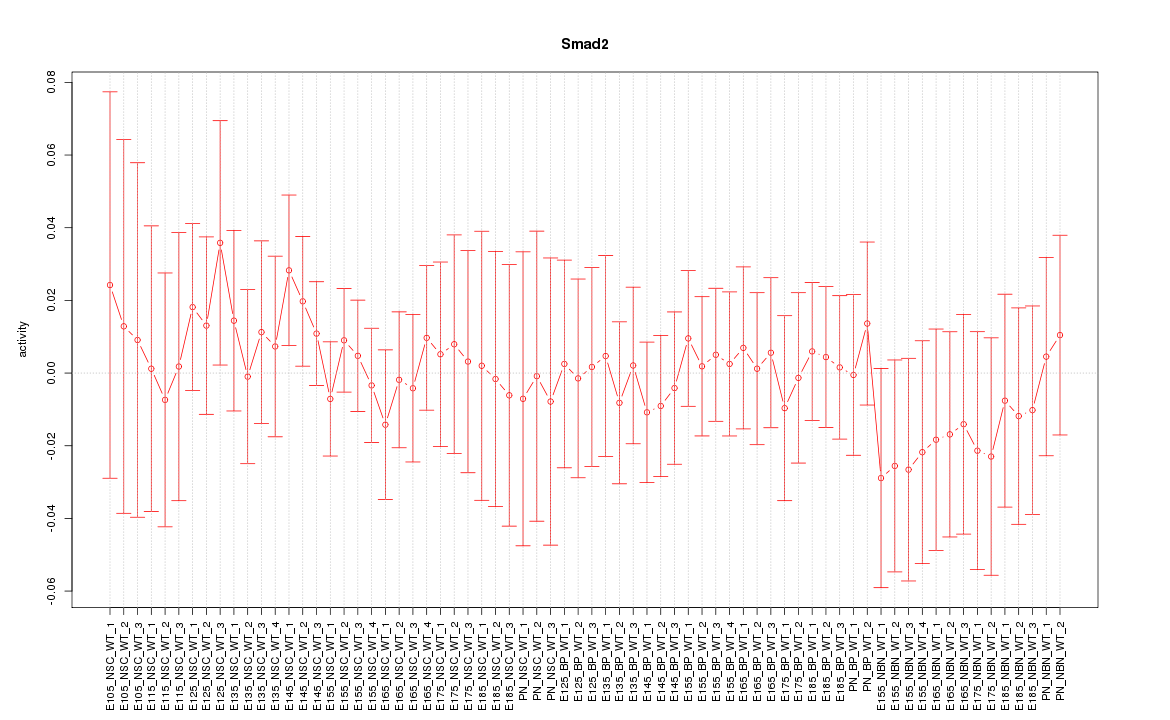
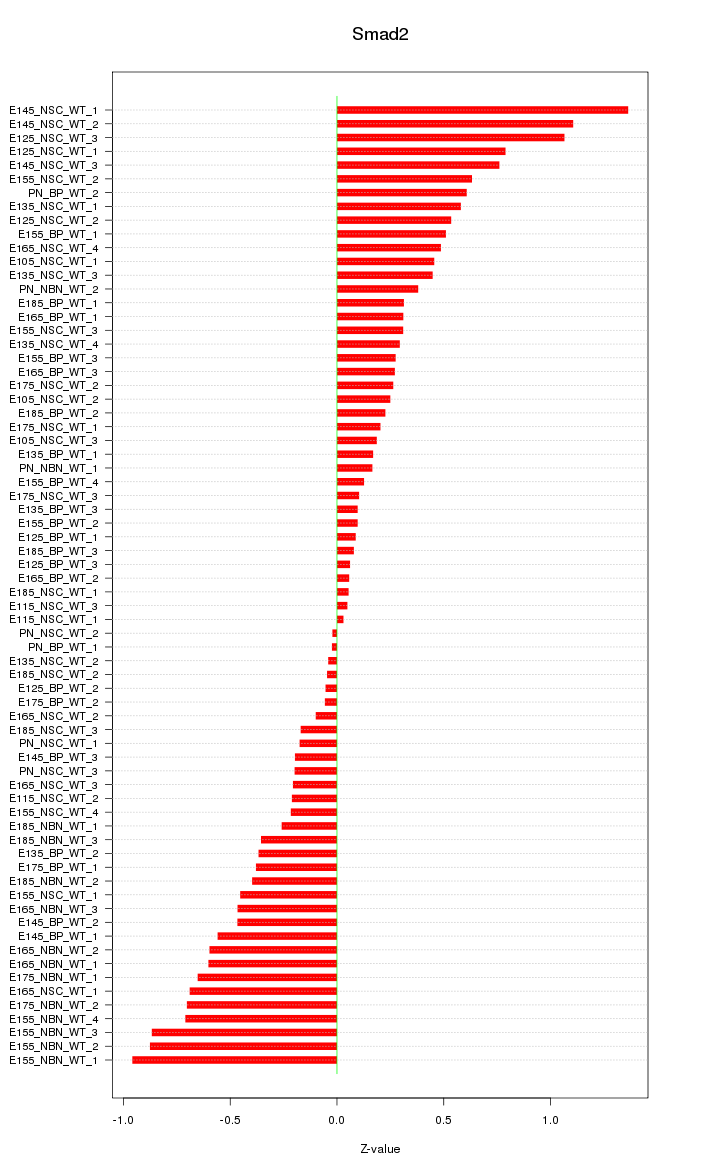
| 1.1 |
3.2 |
GO:0086017 |
response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) Purkinje myocyte action potential(GO:0086017) membrane depolarization during SA node cell action potential(GO:0086046) |
| 1.0 |
2.9 |
GO:0060166 |
olfactory pit development(GO:0060166) |
| 0.7 |
3.4 |
GO:0021764 |
amygdala development(GO:0021764) |
| 0.5 |
3.1 |
GO:0045218 |
zonula adherens maintenance(GO:0045218) |
| 0.4 |
1.1 |
GO:0070844 |
misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.3 |
1.0 |
GO:0061537 |
glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.3 |
1.5 |
GO:1903012 |
positive regulation of bone development(GO:1903012) |
| 0.3 |
1.0 |
GO:0072697 |
protein localization to cell cortex(GO:0072697) |
| 0.3 |
4.2 |
GO:0021796 |
cerebral cortex regionalization(GO:0021796) |
| 0.3 |
1.5 |
GO:0042866 |
pyruvate biosynthetic process(GO:0042866) |
| 0.2 |
0.5 |
GO:1903279 |
regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.1 |
1.0 |
GO:1903300 |
negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 |
0.8 |
GO:0006957 |
complement activation, alternative pathway(GO:0006957) |
| 0.1 |
1.5 |
GO:0061302 |
smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 |
4.0 |
GO:0006270 |
DNA replication initiation(GO:0006270) |
| 0.1 |
1.2 |
GO:0034242 |
negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.1 |
0.5 |
GO:1904936 |
forebrain anterior/posterior pattern specification(GO:0021797) cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 |
0.7 |
GO:0051967 |
negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 |
2.7 |
GO:0030513 |
positive regulation of BMP signaling pathway(GO:0030513) |
| 0.1 |
0.3 |
GO:0017183 |
peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 |
0.9 |
GO:0043248 |
proteasome assembly(GO:0043248) |
| 0.0 |
0.3 |
GO:0035360 |
positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.0 |
0.8 |
GO:0032060 |
bleb assembly(GO:0032060) |
| 0.0 |
1.1 |
GO:0034723 |
DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 |
2.1 |
GO:0035019 |
somatic stem cell population maintenance(GO:0035019) |
| 0.0 |
0.5 |
GO:0060746 |
parental behavior(GO:0060746) |
| 0.0 |
0.2 |
GO:0032020 |
ISG15-protein conjugation(GO:0032020) |
| 0.0 |
0.6 |
GO:0007205 |
protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 |
0.5 |
GO:0007020 |
microtubule nucleation(GO:0007020) |
| 0.0 |
2.2 |
GO:0021954 |
central nervous system neuron development(GO:0021954) |
| 0.0 |
0.5 |
GO:0070979 |
protein K11-linked ubiquitination(GO:0070979) |
| 0.0 |
0.4 |
GO:0000413 |
protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 |
1.6 |
GO:0009636 |
response to toxic substance(GO:0009636) |
| 0.0 |
0.5 |
GO:0070206 |
protein trimerization(GO:0070206) |