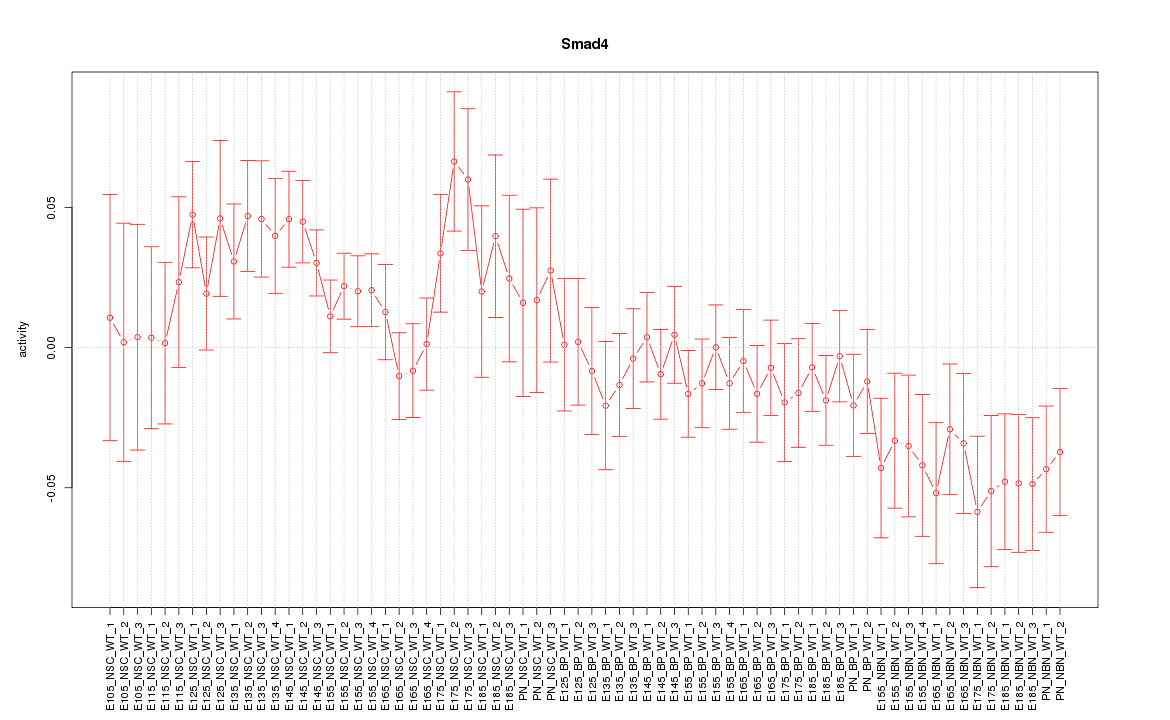
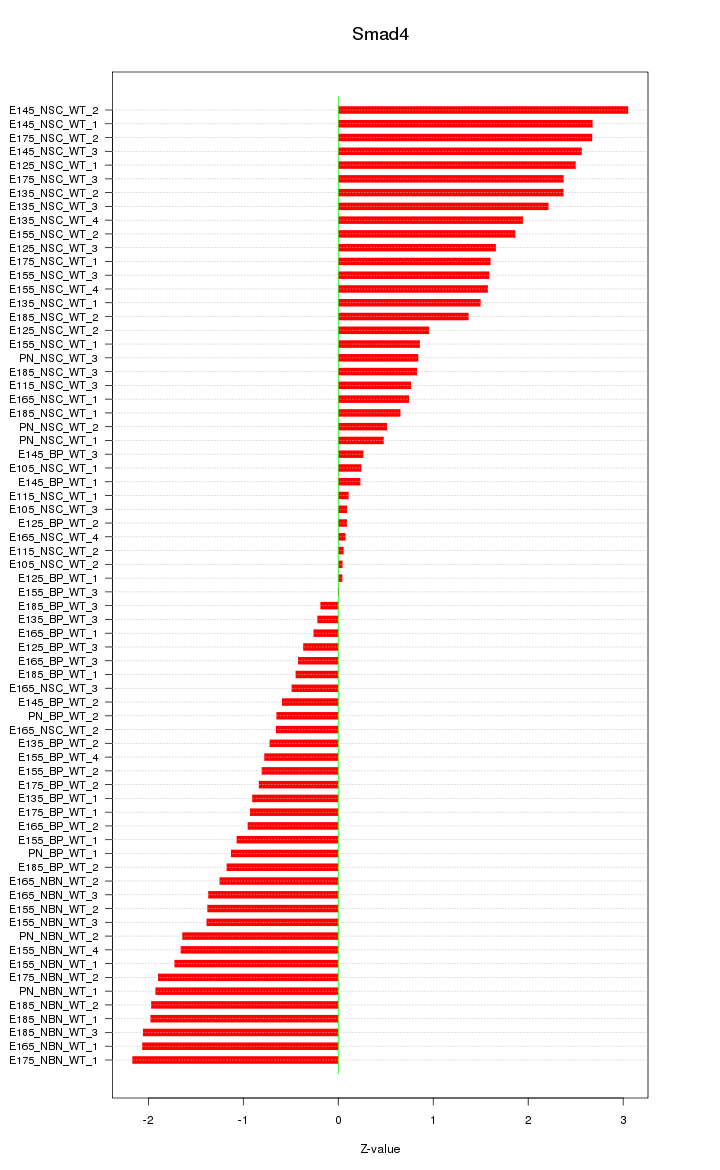
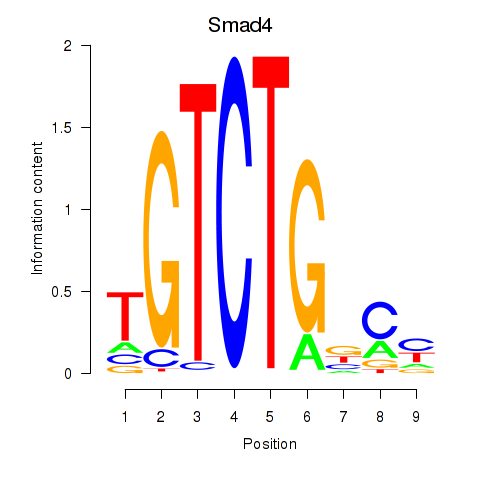
Motif ID: Smad4
Z-value: 1.387
Transcription factors associated with Smad4:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Smad4 | ENSMUSG00000024515.7 | Smad4 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Smad4 | mm10_v2_chr18_-_73703739_73703806 | 0.13 | 2.9e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 12.6 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 4.0 | 27.9 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 4.0 | 11.9 | GO:0015889 | cobalamin transport(GO:0015889) |
| 3.7 | 18.6 | GO:0021764 | amygdala development(GO:0021764) |
| 3.1 | 15.6 | GO:0014719 | skeletal muscle satellite cell activation(GO:0014719) |
| 2.6 | 13.0 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 2.5 | 7.6 | GO:1902460 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 2.5 | 12.4 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 2.1 | 8.4 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 1.7 | 8.6 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 1.6 | 4.9 | GO:0060023 | soft palate development(GO:0060023) |
| 1.4 | 6.9 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 1.3 | 6.3 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) positive regulation of translational initiation by iron(GO:0045994) |
| 1.1 | 21.8 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 1.1 | 5.7 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) cellular response to potassium ion starvation(GO:0051365) |
| 1.1 | 3.4 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 1.1 | 22.2 | GO:0030903 | notochord development(GO:0030903) |
| 1.0 | 5.0 | GO:0060838 | lymphatic endothelial cell fate commitment(GO:0060838) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 1.0 | 2.0 | GO:2000224 | testosterone biosynthetic process(GO:0061370) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.9 | 2.8 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.9 | 2.7 | GO:1901896 | positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.8 | 7.4 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) |
| 0.8 | 6.6 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.8 | 3.0 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.7 | 3.0 | GO:0097709 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) connective tissue replacement(GO:0097709) |
| 0.7 | 15.9 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.7 | 6.7 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.6 | 3.1 | GO:0003223 | ventricular compact myocardium morphogenesis(GO:0003223) proprioception(GO:0019230) |
| 0.6 | 4.3 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.5 | 2.0 | GO:0002339 | B cell selection(GO:0002339) |
| 0.5 | 7.9 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.5 | 3.4 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.4 | 1.3 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.4 | 3.7 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.4 | 3.6 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.4 | 7.7 | GO:0050434 | positive regulation of viral transcription(GO:0050434) |
| 0.4 | 3.3 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.3 | 0.7 | GO:0060681 | branch elongation involved in ureteric bud branching(GO:0060681) |
| 0.3 | 1.3 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 0.3 | 1.0 | GO:0035811 | negative regulation of urine volume(GO:0035811) |
| 0.3 | 1.6 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.3 | 0.9 | GO:0014005 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.3 | 10.1 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.3 | 3.8 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.3 | 4.3 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.3 | 3.2 | GO:0036120 | cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.3 | 3.2 | GO:0001711 | endodermal cell fate commitment(GO:0001711) |
| 0.3 | 3.4 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.3 | 7.7 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.2 | 0.7 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.2 | 2.1 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.2 | 4.3 | GO:0051894 | integrin activation(GO:0033622) positive regulation of focal adhesion assembly(GO:0051894) |
| 0.2 | 0.4 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.2 | 0.8 | GO:0055071 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.2 | 1.8 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.2 | 1.9 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.2 | 2.8 | GO:0031295 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.2 | 0.5 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.1 | 3.2 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 7.6 | GO:0002066 | columnar/cuboidal epithelial cell development(GO:0002066) |
| 0.1 | 2.1 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.1 | 3.5 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 0.6 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 2.1 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 0.5 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 | 4.7 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 2.4 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.1 | 3.0 | GO:0003341 | cilium movement(GO:0003341) |
| 0.1 | 0.7 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 2.0 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.1 | 0.3 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 5.0 | GO:0021954 | central nervous system neuron development(GO:0021954) |
| 0.0 | 0.5 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.0 | 0.1 | GO:0032415 | regulation of sodium:proton antiporter activity(GO:0032415) positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.0 | 1.1 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 2.9 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 3.8 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.7 | GO:0045672 | positive regulation of osteoclast differentiation(GO:0045672) |
| 0.0 | 0.3 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.4 | GO:0060481 | lobar bronchus epithelium development(GO:0060481) |
| 0.0 | 1.8 | GO:1900046 | regulation of blood coagulation(GO:0030193) regulation of hemostasis(GO:1900046) |
| 0.0 | 1.8 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 1.2 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 2.5 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.2 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.0 | 1.2 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 1.5 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 1.4 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 7.7 | GO:0016055 | Wnt signaling pathway(GO:0016055) cell-cell signaling by wnt(GO:0198738) |
| 0.0 | 0.4 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 1.0 | GO:0030855 | epithelial cell differentiation(GO:0030855) |
| 0.0 | 1.2 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 1.2 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 0.9 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.0 | 0.3 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.3 | 15.9 | GO:0038045 | large latent transforming growth factor-beta complex(GO:0038045) |
| 1.1 | 3.4 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.9 | 27.9 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.9 | 2.7 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.8 | 3.0 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.7 | 7.4 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.6 | 13.0 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.5 | 7.0 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.5 | 3.8 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.4 | 3.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.3 | 26.5 | GO:0000786 | nucleosome(GO:0000786) |
| 0.3 | 10.7 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.3 | 6.6 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.3 | 3.5 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.2 | 3.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 0.6 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.2 | 12.4 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.2 | 3.6 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.2 | 2.1 | GO:0005922 | connexon complex(GO:0005922) |
| 0.2 | 2.1 | GO:0032039 | integrator complex(GO:0032039) |
| 0.2 | 10.4 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 1.0 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 5.7 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.1 | 1.5 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 1.7 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 41.4 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 1.3 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 1.6 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 1.1 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 1.0 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 1.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 2.5 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 3.0 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.9 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 21.7 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 1.2 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 8.4 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.4 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.5 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 3.2 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.2 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.1 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.4 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 4.3 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 11.5 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 0.9 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 9.5 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 2.0 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 2.5 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 1.0 | GO:0055037 | recycling endosome(GO:0055037) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 12.6 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 4.1 | 16.3 | GO:0050436 | microfibril binding(GO:0050436) |
| 4.0 | 27.9 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 2.2 | 6.7 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 1.7 | 11.9 | GO:0031419 | cobalamin binding(GO:0031419) |
| 1.1 | 6.3 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 1.0 | 12.4 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.8 | 5.0 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.7 | 10.7 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.7 | 7.7 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.6 | 10.1 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.5 | 3.2 | GO:0043426 | MRF binding(GO:0043426) |
| 0.5 | 2.0 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.5 | 7.7 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.5 | 2.8 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.4 | 11.0 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.4 | 7.6 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.3 | 6.9 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.3 | 3.6 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.3 | 18.6 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.3 | 5.7 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.3 | 0.9 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.3 | 3.6 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.3 | 9.7 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.3 | 2.1 | GO:0031432 | titin binding(GO:0031432) |
| 0.2 | 25.4 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.2 | 8.5 | GO:0070888 | E-box binding(GO:0070888) |
| 0.2 | 3.1 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.2 | 2.0 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.2 | 2.7 | GO:0051861 | glycolipid binding(GO:0051861) |
| 0.2 | 0.8 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.2 | 3.5 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.2 | 2.1 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.2 | 6.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.2 | 1.0 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.2 | 1.8 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 3.3 | GO:0043747 | protein-N-terminal asparagine amidohydrolase activity(GO:0008418) UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase activity(GO:0008759) iprodione amidohydrolase activity(GO:0018748) (3,5-dichlorophenylurea)acetate amidohydrolase activity(GO:0018749) 4'-(2-hydroxyisopropyl)phenylurea amidohydrolase activity(GO:0034571) didemethylisoproturon amidohydrolase activity(GO:0034573) N-isopropylacetanilide amidohydrolase activity(GO:0034576) N-cyclohexylformamide amidohydrolase activity(GO:0034781) isonicotinic acid hydrazide hydrolase activity(GO:0034876) cis-aconitamide amidase activity(GO:0034882) gamma-N-formylaminovinylacetate hydrolase activity(GO:0034885) N2-acetyl-L-lysine deacetylase activity(GO:0043747) O-succinylbenzoate synthase activity(GO:0043748) indoleacetamide hydrolase activity(GO:0043864) N-acetylcitrulline deacetylase activity(GO:0043909) N-acetylgalactosamine-6-phosphate deacetylase activity(GO:0047419) diacetylchitobiose deacetylase activity(GO:0052773) chitooligosaccharide deacetylase activity(GO:0052790) |
| 0.1 | 3.8 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 3.2 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 0.4 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.1 | 4.3 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 8.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 21.8 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 4.8 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.1 | 1.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 3.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.7 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 0.3 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.1 | 1.3 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 7.6 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 11.0 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 1.3 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 4.7 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.3 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 7.7 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 2.3 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 1.6 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.7 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.5 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 4.4 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.3 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 1.1 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.0 | 2.9 | GO:0003735 | structural constituent of ribosome(GO:0003735) |