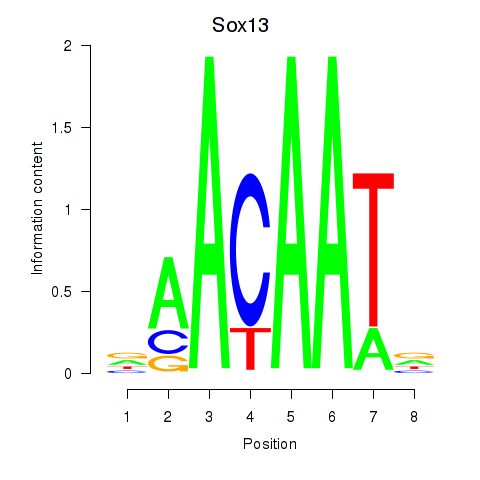
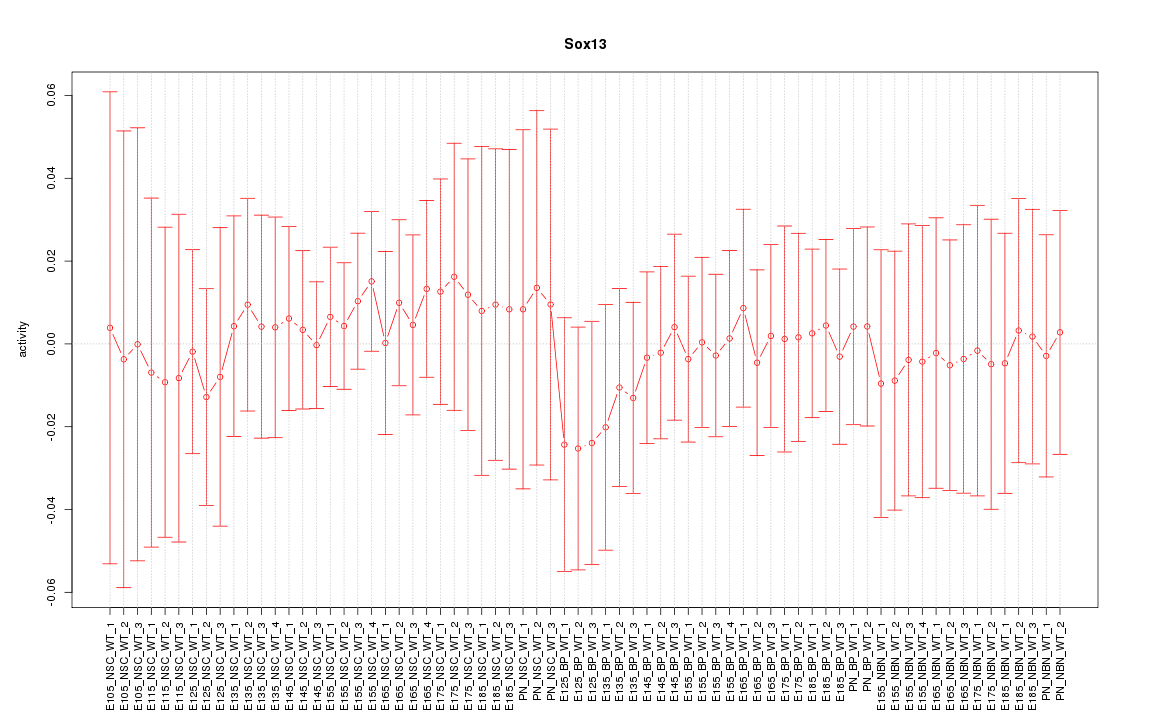
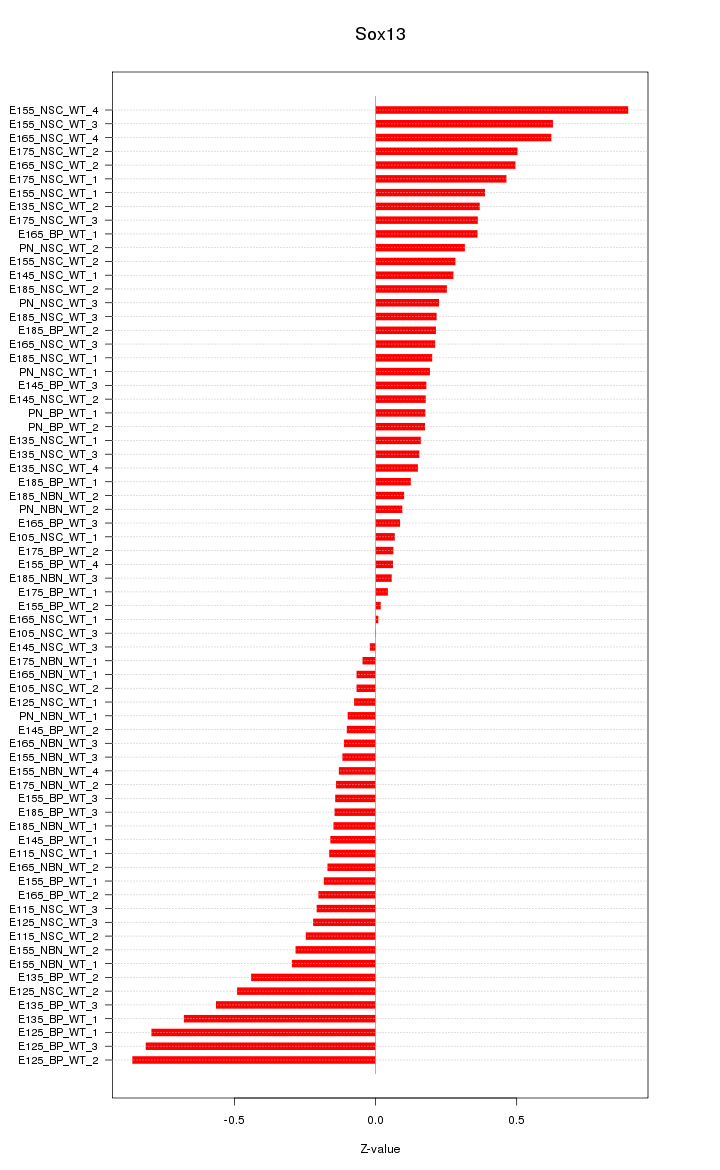
| 0.4 |
2.5 |
GO:0071699 |
olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.2 |
0.9 |
GO:0032289 |
central nervous system myelin formation(GO:0032289) cardiac cell fate specification(GO:0060912) |
| 0.2 |
0.9 |
GO:1904565 |
response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.2 |
2.3 |
GO:0014807 |
regulation of somitogenesis(GO:0014807) |
| 0.2 |
2.8 |
GO:0021684 |
cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.2 |
1.2 |
GO:1904936 |
cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 |
0.5 |
GO:0001550 |
ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 |
0.9 |
GO:0002903 |
negative regulation of B cell apoptotic process(GO:0002903) |
| 0.1 |
0.9 |
GO:0042118 |
endothelial cell activation(GO:0042118) |
| 0.1 |
0.5 |
GO:0046549 |
retinal cone cell development(GO:0046549) |
| 0.1 |
0.3 |
GO:0003192 |
mitral valve formation(GO:0003192) cell migration involved in endocardial cushion formation(GO:0003273) condensed mesenchymal cell proliferation(GO:0072137) |
| 0.0 |
0.4 |
GO:0009186 |
deoxyribonucleoside diphosphate metabolic process(GO:0009186) |
| 0.0 |
0.4 |
GO:0061179 |
negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 |
0.1 |
GO:0060648 |
mammary gland bud morphogenesis(GO:0060648) |
| 0.0 |
0.5 |
GO:1902043 |
positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 |
0.3 |
GO:0050860 |
negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 |
1.4 |
GO:0070527 |
platelet aggregation(GO:0070527) |
| 0.0 |
0.9 |
GO:0030901 |
midbrain development(GO:0030901) |
| 0.0 |
0.2 |
GO:0031937 |
positive regulation of chromatin silencing(GO:0031937) |
| 0.0 |
0.4 |
GO:0034723 |
DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-independent nucleosome assembly(GO:0006336) DNA replication-dependent nucleosome organization(GO:0034723) DNA replication-independent nucleosome organization(GO:0034724) |