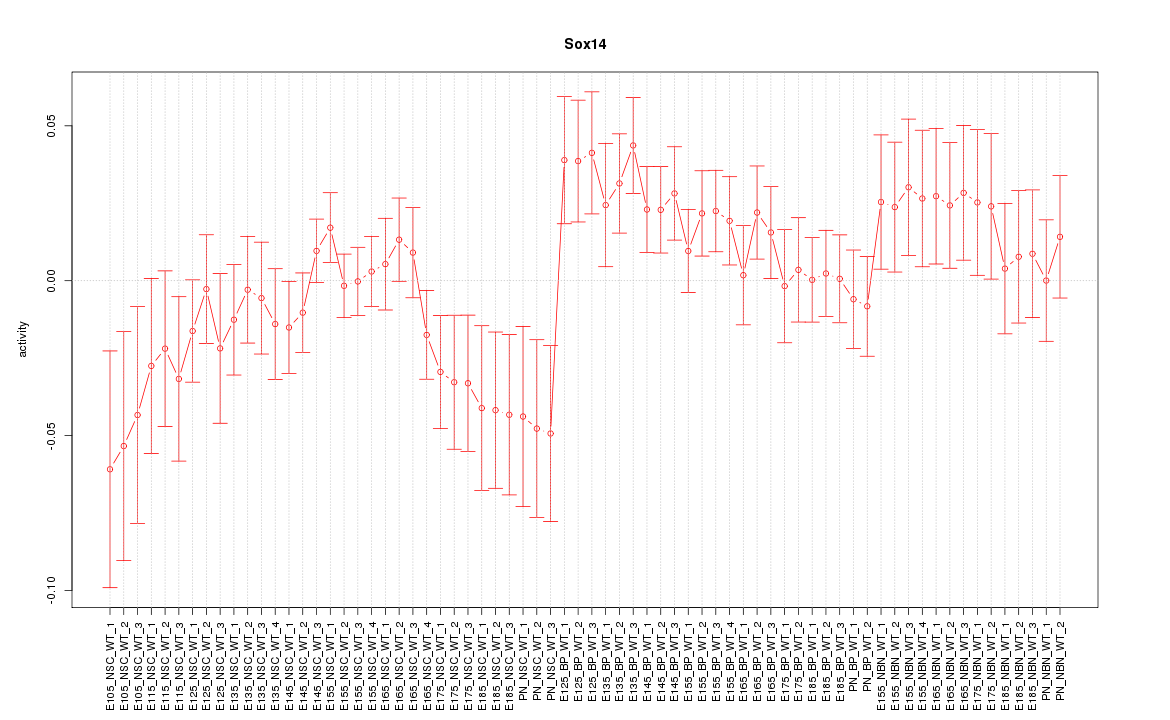
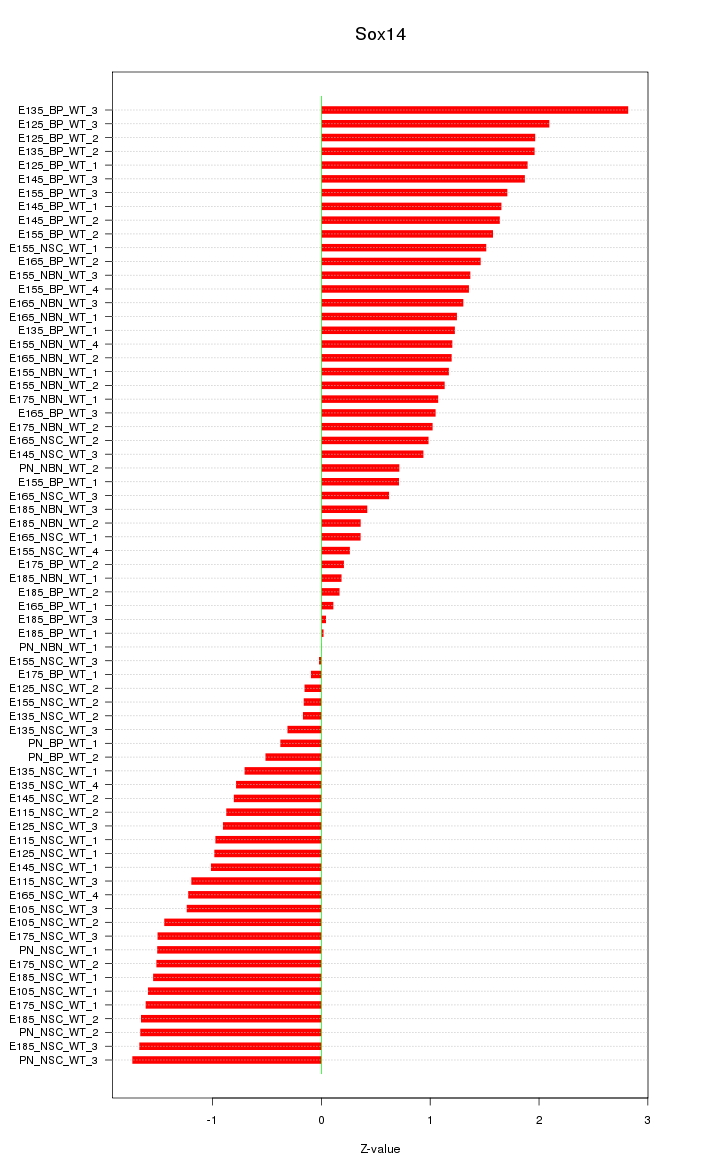
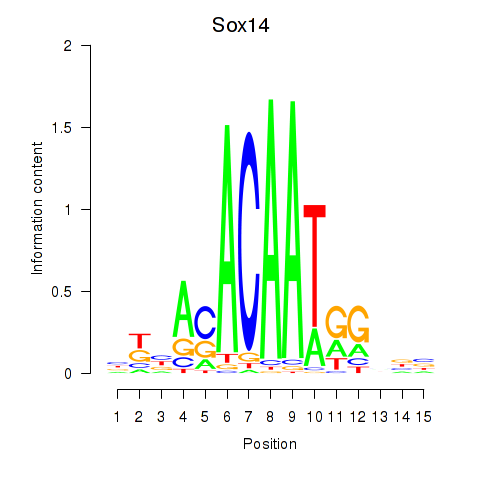
Motif ID: Sox14
Z-value: 1.211
Transcription factors associated with Sox14:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Sox14 | ENSMUSG00000053747.8 | Sox14 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sox14 | mm10_v2_chr9_-_99876147_99876193 | -0.34 | 3.7e-03 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 13.7 | GO:0042560 | 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 4.1 | 16.4 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 2.1 | 6.4 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 2.1 | 8.5 | GO:0032289 | central nervous system myelin formation(GO:0032289) cardiac cell fate specification(GO:0060912) |
| 1.9 | 5.7 | GO:0070428 | regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 1.8 | 5.4 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 1.6 | 6.5 | GO:0036015 | negative regulation of neuron maturation(GO:0014043) response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 1.4 | 4.3 | GO:0003096 | renal sodium ion transport(GO:0003096) |
| 1.4 | 5.5 | GO:1990743 | protein sialylation(GO:1990743) |
| 1.3 | 9.2 | GO:0034047 | regulation of protein phosphatase type 2A activity(GO:0034047) |
| 1.1 | 5.6 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 1.1 | 2.2 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 1.1 | 7.6 | GO:0042518 | negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 1.0 | 6.8 | GO:0061092 | involuntary skeletal muscle contraction(GO:0003011) regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 1.0 | 3.9 | GO:0061428 | embryonic heart tube left/right pattern formation(GO:0060971) negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 1.0 | 11.6 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.9 | 5.6 | GO:0071415 | cellular response to caffeine(GO:0071313) cellular response to purine-containing compound(GO:0071415) |
| 0.9 | 1.7 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.8 | 3.3 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.8 | 9.0 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.8 | 2.4 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.8 | 8.4 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.8 | 2.3 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.7 | 6.6 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.7 | 16.4 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.7 | 2.1 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.7 | 7.7 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.7 | 2.7 | GO:0051582 | positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.7 | 2.0 | GO:0030070 | insulin processing(GO:0030070) |
| 0.6 | 8.4 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.6 | 5.1 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.6 | 8.8 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.6 | 7.4 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.6 | 8.1 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.5 | 3.7 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.5 | 1.6 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.5 | 5.7 | GO:0015697 | quaternary ammonium group transport(GO:0015697) |
| 0.5 | 2.3 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.4 | 2.7 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.4 | 7.1 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.4 | 2.0 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.4 | 1.6 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.4 | 1.6 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.4 | 1.2 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.4 | 2.7 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.4 | 3.1 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.4 | 3.0 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.4 | 7.2 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) |
| 0.4 | 1.5 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.4 | 3.4 | GO:0043615 | astrocyte cell migration(GO:0043615) positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.4 | 1.9 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.4 | 2.6 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.4 | 8.2 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.3 | 2.1 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.3 | 2.4 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.3 | 9.4 | GO:0022401 | desensitization of G-protein coupled receptor protein signaling pathway(GO:0002029) negative adaptation of signaling pathway(GO:0022401) |
| 0.3 | 3.4 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.3 | 1.3 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.3 | 0.6 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.3 | 1.9 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.3 | 5.9 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.3 | 2.2 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.3 | 5.7 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.3 | 3.8 | GO:1901898 | negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.3 | 5.1 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.3 | 1.4 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.3 | 0.3 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.3 | 5.7 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.3 | 1.0 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.3 | 7.1 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.2 | 6.1 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 13.3 | GO:0032410 | negative regulation of transporter activity(GO:0032410) |
| 0.2 | 1.9 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.2 | 0.7 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.2 | 4.5 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.2 | 1.1 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.2 | 1.1 | GO:0050912 | detection of chemical stimulus involved in sensory perception(GO:0050907) detection of chemical stimulus involved in sensory perception of taste(GO:0050912) |
| 0.2 | 0.9 | GO:0090238 | positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.2 | 1.6 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.2 | 4.3 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.2 | 29.9 | GO:0051017 | actin filament bundle assembly(GO:0051017) |
| 0.2 | 1.9 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.2 | 0.6 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.2 | 1.3 | GO:0051461 | protein import into peroxisome matrix, docking(GO:0016560) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.2 | 3.1 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.2 | 8.0 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.2 | 1.0 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.2 | 3.2 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.2 | 0.7 | GO:0006867 | asparagine transport(GO:0006867) glutamine transport(GO:0006868) |
| 0.2 | 8.1 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.2 | 2.1 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.2 | 0.5 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.2 | 0.6 | GO:0034145 | positive regulation of toll-like receptor 4 signaling pathway(GO:0034145) |
| 0.1 | 1.2 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.1 | 4.3 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 9.2 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.1 | 0.5 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.1 | 6.9 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.1 | 19.3 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.1 | 4.6 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.1 | 1.5 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 1.0 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.1 | 5.0 | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.1 | 0.5 | GO:1904690 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.1 | 3.3 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.1 | 6.2 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.1 | 1.0 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 | 0.3 | GO:0009193 | pyrimidine ribonucleoside diphosphate metabolic process(GO:0009193) UDP metabolic process(GO:0046048) |
| 0.1 | 5.2 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.1 | 3.0 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.1 | 2.9 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.1 | 3.0 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.1 | 2.4 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.1 | 1.8 | GO:0006221 | pyrimidine nucleotide biosynthetic process(GO:0006221) |
| 0.1 | 1.5 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.1 | 0.8 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 2.1 | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0070059) |
| 0.1 | 2.1 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 1.0 | GO:0015858 | nucleoside transport(GO:0015858) |
| 0.0 | 2.2 | GO:0030317 | sperm motility(GO:0030317) |
| 0.0 | 0.4 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 1.1 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 6.9 | GO:0006906 | vesicle fusion(GO:0006906) |
| 0.0 | 0.4 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 4.3 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 0.4 | GO:1901409 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 4.5 | GO:0098792 | xenophagy(GO:0098792) |
| 0.0 | 2.1 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.0 | 0.8 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.2 | GO:0090241 | negative regulation of histone H4 acetylation(GO:0090241) |
| 0.0 | 0.7 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 3.8 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 0.4 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.0 | 2.0 | GO:0030705 | cytoskeleton-dependent intracellular transport(GO:0030705) |
| 0.0 | 0.8 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.0 | 0.8 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 5.5 | GO:0007409 | axonogenesis(GO:0007409) |
| 0.0 | 0.4 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.5 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 2.1 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 2.4 | GO:0001764 | neuron migration(GO:0001764) |
| 0.0 | 2.5 | GO:0007626 | locomotory behavior(GO:0007626) |
| 0.0 | 0.6 | GO:1901799 | negative regulation of proteasomal protein catabolic process(GO:1901799) negative regulation of proteolysis involved in cellular protein catabolic process(GO:1903051) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 16.4 | GO:0043259 | laminin-1 complex(GO:0005606) laminin-10 complex(GO:0043259) |
| 1.4 | 8.4 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 1.1 | 5.6 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.8 | 2.5 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.7 | 0.7 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.6 | 6.5 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.6 | 2.9 | GO:1990761 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.5 | 5.5 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.5 | 5.9 | GO:0031258 | catenin complex(GO:0016342) lamellipodium membrane(GO:0031258) |
| 0.5 | 6.6 | GO:0070938 | contractile ring(GO:0070938) |
| 0.5 | 9.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.5 | 3.4 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.4 | 2.6 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.4 | 4.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.4 | 3.2 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.4 | 2.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.4 | 4.6 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.4 | 2.5 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.3 | 2.7 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.3 | 1.3 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.3 | 7.6 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.3 | 7.1 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.2 | 2.6 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.2 | 33.4 | GO:0005884 | actin filament(GO:0005884) |
| 0.2 | 7.7 | GO:0043034 | costamere(GO:0043034) |
| 0.2 | 9.5 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.2 | 1.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.2 | 4.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.2 | 2.4 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.2 | 0.5 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 8.2 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 2.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 5.7 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 2.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 0.5 | GO:0001652 | granular component(GO:0001652) |
| 0.1 | 8.8 | GO:0042641 | actomyosin(GO:0042641) |
| 0.1 | 0.8 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.1 | 10.2 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 1.2 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.1 | 6.4 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.1 | 23.0 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.1 | 11.8 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 1.1 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 10.2 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 1.9 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 4.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 0.9 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 0.4 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 11.3 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 1.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 1.5 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.4 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 3.1 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 4.5 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 4.4 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.0 | 8.2 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 1.3 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 1.1 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.1 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 0.6 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 1.8 | GO:0005768 | endosome(GO:0005768) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 13.7 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 2.3 | 9.3 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 2.0 | 16.4 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 2.0 | 10.0 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 1.9 | 5.6 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 1.5 | 7.6 | GO:0052833 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 1.4 | 8.2 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 1.2 | 6.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 1.1 | 7.7 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.8 | 8.4 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.8 | 3.3 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.8 | 13.8 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.8 | 5.7 | GO:0015651 | quaternary ammonium group transmembrane transporter activity(GO:0015651) |
| 0.7 | 2.1 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 0.7 | 9.2 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.5 | 1.6 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.5 | 1.6 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.5 | 1.6 | GO:0070890 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.5 | 11.6 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.5 | 6.1 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.5 | 3.9 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.5 | 1.9 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.4 | 1.8 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.4 | 1.7 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.4 | 5.0 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.4 | 5.7 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.4 | 8.1 | GO:0043747 | protein-N-terminal asparagine amidohydrolase activity(GO:0008418) UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase activity(GO:0008759) iprodione amidohydrolase activity(GO:0018748) (3,5-dichlorophenylurea)acetate amidohydrolase activity(GO:0018749) 4'-(2-hydroxyisopropyl)phenylurea amidohydrolase activity(GO:0034571) didemethylisoproturon amidohydrolase activity(GO:0034573) N-isopropylacetanilide amidohydrolase activity(GO:0034576) N-cyclohexylformamide amidohydrolase activity(GO:0034781) isonicotinic acid hydrazide hydrolase activity(GO:0034876) cis-aconitamide amidase activity(GO:0034882) gamma-N-formylaminovinylacetate hydrolase activity(GO:0034885) N2-acetyl-L-lysine deacetylase activity(GO:0043747) O-succinylbenzoate synthase activity(GO:0043748) indoleacetamide hydrolase activity(GO:0043864) N-acetylcitrulline deacetylase activity(GO:0043909) N-acetylgalactosamine-6-phosphate deacetylase activity(GO:0047419) diacetylchitobiose deacetylase activity(GO:0052773) chitooligosaccharide deacetylase activity(GO:0052790) |
| 0.4 | 1.1 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.4 | 1.4 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.3 | 6.8 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.3 | 9.4 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.3 | 2.2 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.3 | 9.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.3 | 2.4 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.3 | 5.7 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.3 | 4.1 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.3 | 2.4 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.3 | 1.9 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.3 | 10.4 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.3 | 1.3 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.2 | 5.9 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.2 | 2.7 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.2 | 6.2 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.2 | 4.1 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.2 | 2.3 | GO:0052872 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) phenanthrene 9,10-monooxygenase activity(GO:0018636) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) tocotrienol omega-hydroxylase activity(GO:0052872) thalianol hydroxylase activity(GO:0080014) |
| 0.2 | 3.3 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.2 | 3.7 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.2 | 0.6 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.2 | 7.1 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.2 | 2.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.2 | 2.1 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.2 | 29.9 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.2 | 1.6 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.2 | 1.0 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.2 | 2.9 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.2 | 3.9 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.2 | 2.9 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.2 | 1.0 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.2 | 8.6 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.2 | 2.6 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.2 | 0.5 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.2 | 0.8 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.2 | 5.7 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 1.2 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.1 | 2.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 5.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 7.5 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.7 | GO:0015186 | L-asparagine transmembrane transporter activity(GO:0015182) L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 14.3 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 1.6 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 38.0 | GO:0043774 | UDP-N-acetylmuramoylalanyl-D-glutamyl-2,6-diaminopimelate-D-alanyl-D-alanine ligase activity(GO:0008766) ribosomal S6-glutamic acid ligase activity(GO:0018169) coenzyme F420-0 gamma-glutamyl ligase activity(GO:0043773) coenzyme F420-2 alpha-glutamyl ligase activity(GO:0043774) protein-glycine ligase activity(GO:0070735) protein-glycine ligase activity, initiating(GO:0070736) protein-glycine ligase activity, elongating(GO:0070737) tubulin-glycine ligase activity(GO:0070738) |
| 0.1 | 3.8 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 0.9 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.1 | 1.4 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 2.9 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 5.0 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 0.4 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 1.9 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 1.2 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 0.6 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 1.9 | GO:0034930 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) heparan sulfate 2-O-sulfotransferase activity(GO:0004394) HNK-1 sulfotransferase activity(GO:0016232) heparan sulfate 6-O-sulfotransferase activity(GO:0017095) trans-9R,10R-dihydrodiolphenanthrene sulfotransferase activity(GO:0018721) 1-phenanthrol sulfotransferase activity(GO:0018722) 3-phenanthrol sulfotransferase activity(GO:0018723) 4-phenanthrol sulfotransferase activity(GO:0018724) trans-3,4-dihydrodiolphenanthrene sulfotransferase activity(GO:0018725) 9-phenanthrol sulfotransferase activity(GO:0018726) 2-phenanthrol sulfotransferase activity(GO:0018727) phenanthrol sulfotransferase activity(GO:0019111) 1-hydroxypyrene sulfotransferase activity(GO:0034930) proteoglycan sulfotransferase activity(GO:0050698) cholesterol sulfotransferase activity(GO:0051922) hydroxyjasmonate sulfotransferase activity(GO:0080131) |
| 0.1 | 2.5 | GO:0018602 | sulfonate dioxygenase activity(GO:0000907) 2,4-dichlorophenoxyacetate alpha-ketoglutarate dioxygenase activity(GO:0018602) hypophosphite dioxygenase activity(GO:0034792) gibberellin 2-beta-dioxygenase activity(GO:0045543) C-19 gibberellin 2-beta-dioxygenase activity(GO:0052634) C-20 gibberellin 2-beta-dioxygenase activity(GO:0052635) |
| 0.1 | 1.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 1.0 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.1 | 1.0 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.1 | 0.5 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.7 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 1.2 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.6 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.5 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 1.4 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.3 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 1.2 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 8.2 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 4.2 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 1.5 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 2.6 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 2.7 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 2.2 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 1.9 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.4 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.8 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.9 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 2.6 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 0.4 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.5 | GO:0002039 | p53 binding(GO:0002039) |