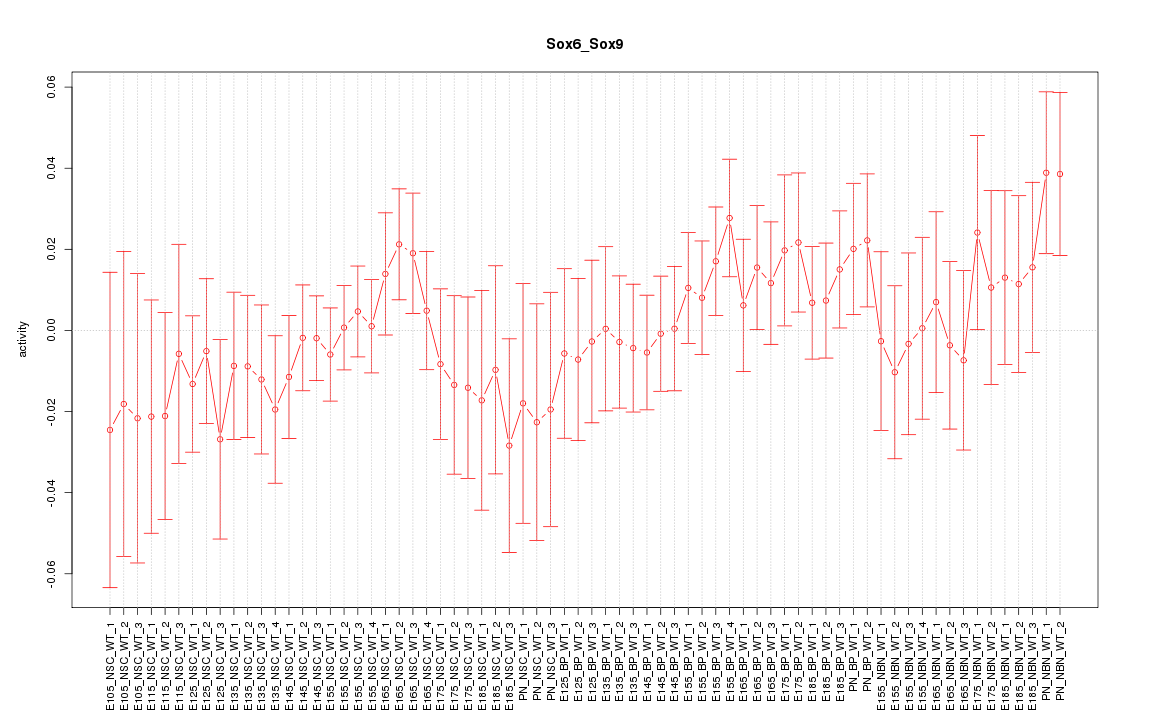
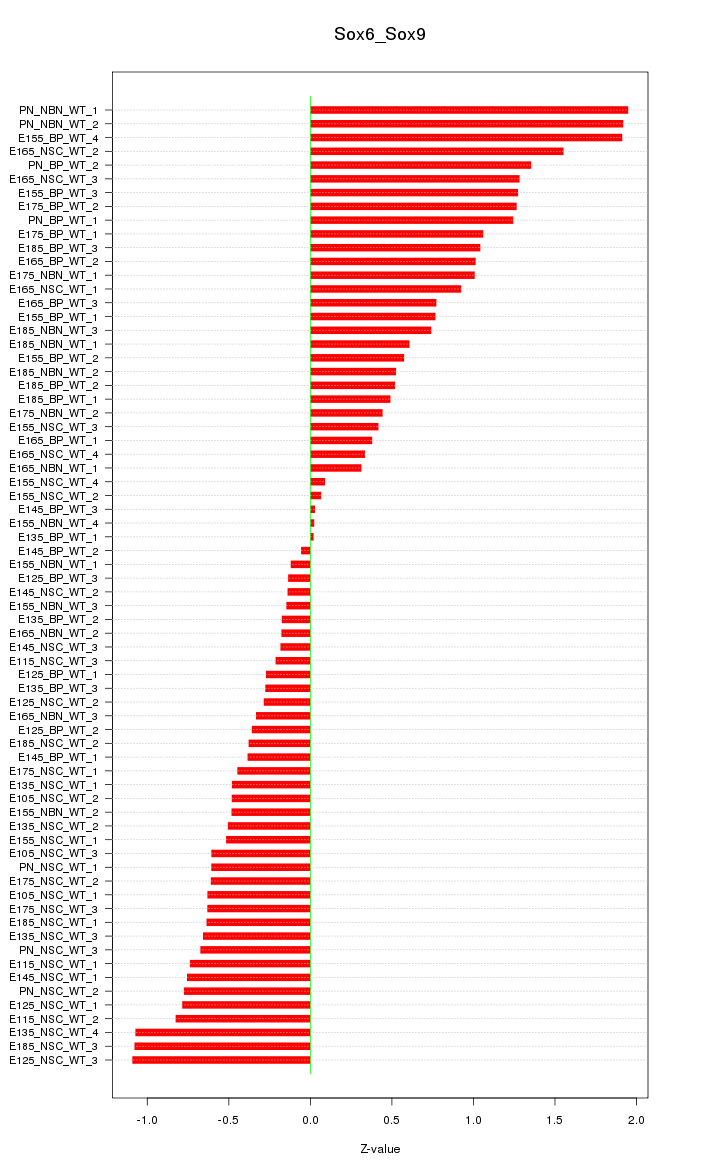
Motif ID: Sox6_Sox9
Z-value: 0.784
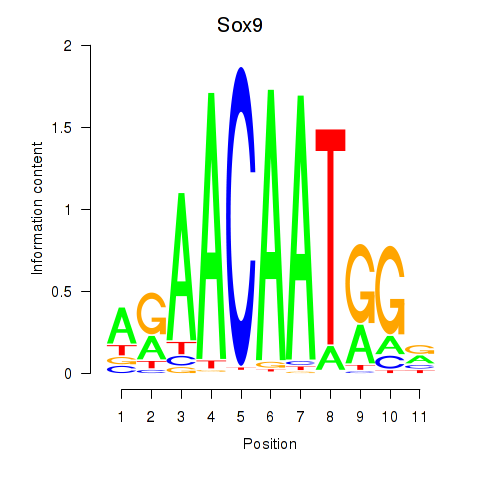
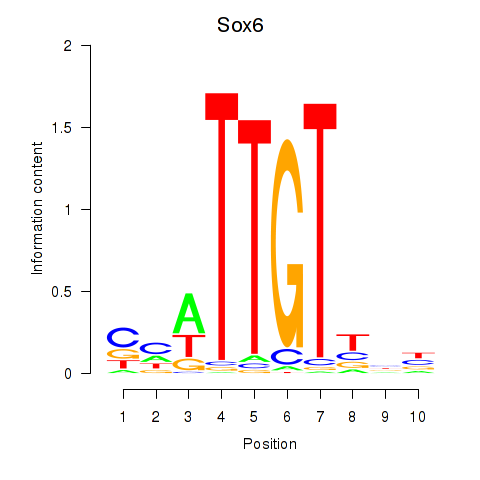
Transcription factors associated with Sox6_Sox9:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Sox6 | ENSMUSG00000051910.7 | Sox6 |
| Sox9 | ENSMUSG00000000567.5 | Sox9 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sox9 | mm10_v2_chr11_+_112782182_112782248 | -0.58 | 1.1e-07 | Click! |
| Sox6 | mm10_v2_chr7_-_116038734_116038750 | -0.41 | 4.9e-04 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.4 | 13.3 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 1.9 | 7.7 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 1.8 | 5.3 | GO:0042560 | 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 1.6 | 4.8 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 1.2 | 4.8 | GO:0032289 | central nervous system myelin formation(GO:0032289) cardiac cell fate specification(GO:0060912) |
| 1.2 | 10.5 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 1.0 | 5.1 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.9 | 3.7 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.9 | 9.1 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.8 | 5.7 | GO:0034047 | regulation of protein phosphatase type 2A activity(GO:0034047) |
| 0.7 | 2.1 | GO:0043379 | memory T cell differentiation(GO:0043379) negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.7 | 4.1 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.6 | 2.6 | GO:1990743 | protein sialylation(GO:1990743) |
| 0.6 | 2.5 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.6 | 8.6 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.6 | 3.4 | GO:0021814 | cell motility involved in cerebral cortex radial glia guided migration(GO:0021814) |
| 0.6 | 6.6 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.5 | 4.3 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.5 | 2.1 | GO:0003357 | noradrenergic neuron differentiation(GO:0003357) |
| 0.5 | 2.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.5 | 2.0 | GO:0086042 | cardiac muscle cell-cardiac muscle cell adhesion(GO:0086042) |
| 0.5 | 2.4 | GO:0070305 | response to cGMP(GO:0070305) cellular response to cGMP(GO:0071321) |
| 0.5 | 7.8 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.5 | 1.4 | GO:2000616 | negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 0.5 | 1.4 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.4 | 2.2 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.4 | 1.3 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.4 | 2.9 | GO:0042518 | negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.4 | 3.1 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) |
| 0.4 | 4.2 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.4 | 1.9 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.4 | 1.5 | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 0.4 | 1.5 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.3 | 6.6 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.3 | 1.3 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.3 | 5.6 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.3 | 3.3 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.3 | 0.9 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.3 | 0.9 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) |
| 0.3 | 1.4 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.3 | 1.9 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.3 | 3.0 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.3 | 2.1 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.3 | 1.0 | GO:0032512 | regulation of protein phosphatase type 2B activity(GO:0032512) positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.2 | 6.0 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 2.3 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.2 | 0.7 | GO:0090325 | regulation of locomotion involved in locomotory behavior(GO:0090325) negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.2 | 3.3 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.2 | 1.6 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.2 | 1.1 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.2 | 2.4 | GO:0048505 | regulation of timing of cell differentiation(GO:0048505) |
| 0.2 | 2.7 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.2 | 1.8 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.2 | 0.6 | GO:1902524 | positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 0.2 | 5.0 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.2 | 0.6 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.2 | 0.6 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.2 | 1.2 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.2 | 3.5 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.2 | 1.9 | GO:0007379 | segment specification(GO:0007379) |
| 0.2 | 1.3 | GO:0046639 | negative regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043371) negative regulation of T-helper cell differentiation(GO:0045623) negative regulation of alpha-beta T cell differentiation(GO:0046639) |
| 0.2 | 2.0 | GO:0051923 | sulfation(GO:0051923) |
| 0.2 | 1.5 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.2 | 2.0 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.2 | 1.2 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.2 | 1.0 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.2 | 1.5 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.2 | 1.5 | GO:0098907 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) regulation of SA node cell action potential(GO:0098907) |
| 0.2 | 0.8 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.2 | 4.7 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 0.3 | GO:2000790 | regulation of mesenchymal cell proliferation involved in lung development(GO:2000790) negative regulation of mesenchymal cell proliferation involved in lung development(GO:2000791) |
| 0.1 | 0.7 | GO:0071476 | cellular hypotonic response(GO:0071476) |
| 0.1 | 0.9 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.1 | 0.7 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.1 | 1.0 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.1 | 0.4 | GO:0071544 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.1 | 0.7 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.1 | 0.4 | GO:0060743 | estrous cycle(GO:0044849) epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.1 | 3.2 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.1 | 0.8 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.1 | 3.7 | GO:0010043 | response to zinc ion(GO:0010043) |
| 0.1 | 0.9 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.1 | 1.8 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.1 | 0.8 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.1 | 0.5 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.1 | 0.2 | GO:0002741 | positive regulation of cytokine secretion involved in immune response(GO:0002741) |
| 0.1 | 7.4 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 0.4 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.1 | 0.5 | GO:0072362 | regulation of glycolytic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072362) |
| 0.1 | 0.7 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.6 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 0.8 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 0.4 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 0.5 | GO:0050912 | detection of chemical stimulus involved in sensory perception(GO:0050907) detection of chemical stimulus involved in sensory perception of taste(GO:0050912) |
| 0.1 | 1.0 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.1 | 1.8 | GO:0048305 | immunoglobulin secretion(GO:0048305) |
| 0.1 | 0.9 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.1 | 0.6 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.1 | 0.5 | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.1 | 0.6 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 | 0.3 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.1 | 0.2 | GO:0000965 | mitochondrial RNA 3'-end processing(GO:0000965) |
| 0.1 | 2.2 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 0.7 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.1 | 0.5 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.1 | 1.4 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.1 | 0.3 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.1 | 1.2 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.1 | 0.9 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.1 | 0.1 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 0.1 | 1.9 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.1 | 3.8 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.1 | 0.3 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.1 | 4.2 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 0.4 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.3 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.1 | 0.5 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 0.4 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.9 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 3.3 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.1 | 0.4 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.1 | 0.6 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 | 2.1 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.3 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 | 7.7 | GO:0030041 | actin filament polymerization(GO:0030041) |
| 0.0 | 0.7 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 1.3 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.0 | 1.2 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 0.2 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.9 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.0 | 7.0 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 0.8 | GO:0005980 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.7 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.1 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.0 | 0.5 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 | 5.1 | GO:0030705 | cytoskeleton-dependent intracellular transport(GO:0030705) |
| 0.0 | 3.5 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.6 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 2.3 | GO:0030010 | establishment of cell polarity(GO:0030010) |
| 0.0 | 0.6 | GO:0045026 | plasma membrane fusion(GO:0045026) |
| 0.0 | 0.3 | GO:0033008 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 4.0 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 1.2 | GO:0030517 | negative regulation of axon extension(GO:0030517) |
| 0.0 | 0.4 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.4 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.4 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.0 | 1.2 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 2.0 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 0.6 | GO:0045921 | positive regulation of exocytosis(GO:0045921) |
| 0.0 | 0.1 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.3 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 4.3 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.0 | 0.5 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.3 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.4 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.1 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.9 | GO:0010977 | negative regulation of neuron projection development(GO:0010977) |
| 0.0 | 2.0 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 9.1 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 2.0 | 7.8 | GO:0044307 | dendritic branch(GO:0044307) |
| 1.9 | 7.7 | GO:0043259 | laminin-1 complex(GO:0005606) laminin-10 complex(GO:0043259) |
| 0.6 | 2.4 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.5 | 1.4 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.4 | 3.8 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.4 | 1.5 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.4 | 1.9 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.3 | 5.1 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.3 | 2.5 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.3 | 1.2 | GO:0031673 | H zone(GO:0031673) |
| 0.3 | 5.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.3 | 0.9 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.3 | 0.8 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.3 | 2.6 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.2 | 4.2 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.2 | 1.0 | GO:0000798 | nuclear cohesin complex(GO:0000798) nuclear meiotic cohesin complex(GO:0034991) |
| 0.2 | 1.7 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.2 | 0.6 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.2 | 1.9 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 8.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 2.9 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.1 | 0.6 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 0.9 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 0.9 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 0.4 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 0.5 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 3.3 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 2.0 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 1.3 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 0.9 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 3.5 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 2.4 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 1.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 1.5 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 6.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 1.5 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 6.5 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 1.0 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 1.7 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 1.9 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 2.0 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.1 | 1.0 | GO:0005640 | nuclear outer membrane(GO:0005640) sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 0.6 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 5.4 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 0.7 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 2.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.5 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.6 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.3 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 3.7 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 4.2 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.6 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 4.1 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.6 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 2.1 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 1.3 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.2 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 9.0 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 2.0 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 5.5 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.8 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.4 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 0.4 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 2.5 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 3.4 | GO:0005773 | vacuole(GO:0005773) |
| 0.0 | 0.8 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 0.2 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 0.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.5 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 3.1 | GO:0005740 | mitochondrial envelope(GO:0005740) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 8.0 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 1.8 | 9.2 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 1.8 | 5.3 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 1.4 | 4.1 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 1.0 | 7.7 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.8 | 12.6 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.8 | 5.4 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.7 | 2.0 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.6 | 3.1 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.6 | 2.4 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.6 | 2.9 | GO:0052833 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.6 | 4.0 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.5 | 2.4 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.5 | 1.4 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.4 | 1.3 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 0.4 | 8.8 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.4 | 3.3 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.4 | 5.7 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.4 | 2.0 | GO:0086080 | connexin binding(GO:0071253) protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.4 | 1.9 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.4 | 1.5 | GO:0031721 | haptoglobin binding(GO:0031720) hemoglobin alpha binding(GO:0031721) |
| 0.3 | 2.3 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.3 | 1.7 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.3 | 1.6 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.3 | 2.7 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.3 | 3.2 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.3 | 4.3 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.3 | 5.1 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.3 | 9.1 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.2 | 2.7 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.2 | 1.0 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.2 | 0.7 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.2 | 5.6 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.2 | 1.5 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.2 | 0.9 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.2 | 1.7 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.2 | 1.4 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.2 | 0.8 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.2 | 5.7 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.2 | 2.2 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.2 | 2.2 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.2 | 1.3 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.2 | 2.1 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.2 | 0.5 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.2 | 2.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.9 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 8.3 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 0.4 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.1 | 0.1 | GO:0031755 | endothelial differentiation G-protein coupled receptor binding(GO:0031753) Edg-2 lysophosphatidic acid receptor binding(GO:0031755) |
| 0.1 | 0.6 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.7 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 3.8 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 1.5 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 2.2 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.9 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 6.0 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 1.5 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.1 | 0.7 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.7 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.4 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.1 | 4.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.4 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.1 | 0.9 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 0.6 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.1 | 0.5 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.1 | 0.5 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.1 | 1.1 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 1.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.7 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 1.0 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.1 | 0.2 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.9 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 2.4 | GO:0044824 | integrase activity(GO:0008907) T/G mismatch-specific endonuclease activity(GO:0043765) retroviral integrase activity(GO:0044823) retroviral 3' processing activity(GO:0044824) |
| 0.0 | 0.9 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 3.7 | GO:0005267 | potassium channel activity(GO:0005267) |
| 0.0 | 0.4 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.7 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 7.4 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.9 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.9 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 8.5 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.5 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 2.0 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 3.8 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 5.5 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 3.7 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.9 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.4 | GO:0019531 | bicarbonate transmembrane transporter activity(GO:0015106) oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 1.9 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.7 | GO:0031690 | adrenergic receptor binding(GO:0031690) |
| 0.0 | 0.5 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.2 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.6 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.3 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.0 | 1.0 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.4 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.8 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.7 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 0.7 | GO:0018602 | sulfonate dioxygenase activity(GO:0000907) 2,4-dichlorophenoxyacetate alpha-ketoglutarate dioxygenase activity(GO:0018602) hypophosphite dioxygenase activity(GO:0034792) gibberellin 2-beta-dioxygenase activity(GO:0045543) C-19 gibberellin 2-beta-dioxygenase activity(GO:0052634) C-20 gibberellin 2-beta-dioxygenase activity(GO:0052635) |
| 0.0 | 5.4 | GO:0043774 | UDP-N-acetylmuramoylalanyl-D-glutamyl-2,6-diaminopimelate-D-alanyl-D-alanine ligase activity(GO:0008766) ribosomal S6-glutamic acid ligase activity(GO:0018169) coenzyme F420-0 gamma-glutamyl ligase activity(GO:0043773) coenzyme F420-2 alpha-glutamyl ligase activity(GO:0043774) protein-glycine ligase activity(GO:0070735) protein-glycine ligase activity, initiating(GO:0070736) protein-glycine ligase activity, elongating(GO:0070737) tubulin-glycine ligase activity(GO:0070738) protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 2.2 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.0 | 0.8 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.4 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 1.0 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.1 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.9 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 5.0 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 0.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.2 | GO:0004549 | tRNA-specific ribonuclease activity(GO:0004549) |
| 0.0 | 0.7 | GO:0005516 | calmodulin binding(GO:0005516) |