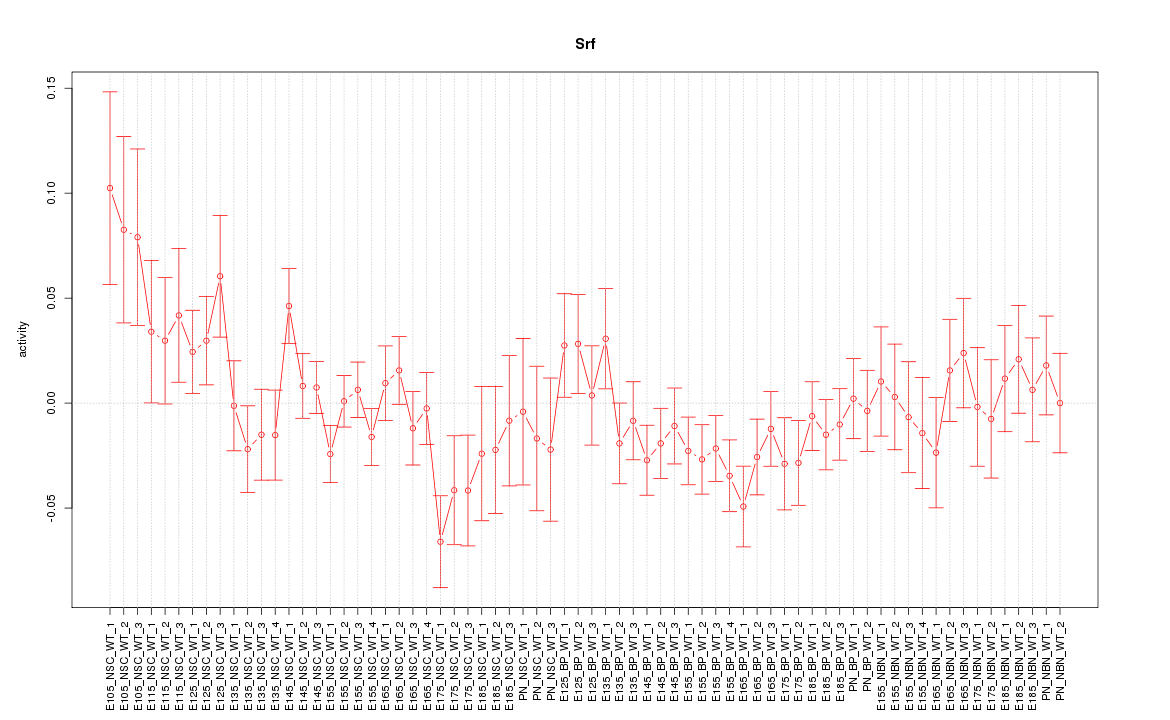
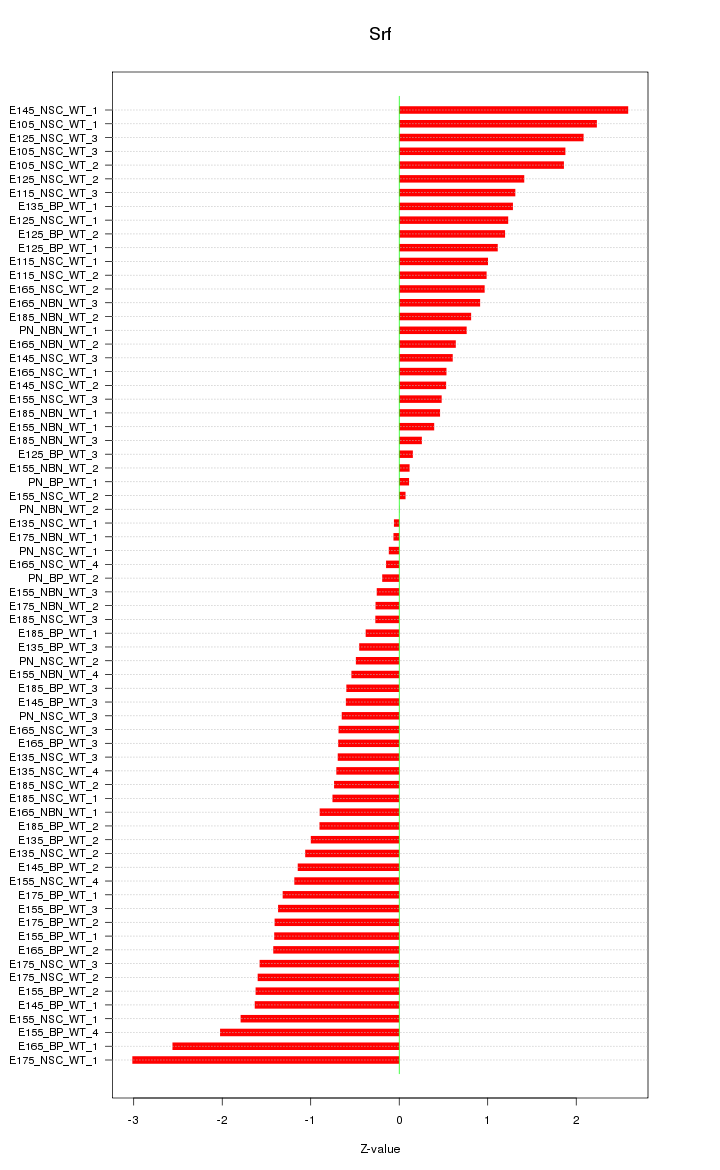
Motif ID: Srf
Z-value: 1.161
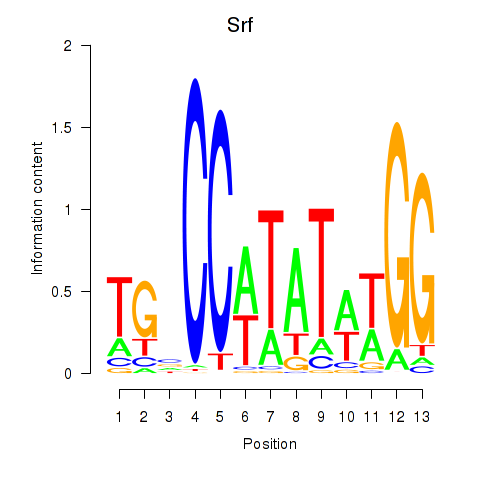
Transcription factors associated with Srf:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Srf | ENSMUSG00000015605.5 | Srf |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Srf | mm10_v2_chr17_-_46556158_46556188 | -0.10 | 4.1e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 18.6 | GO:0030091 | protein repair(GO:0030091) |
| 3.5 | 20.9 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 1.6 | 4.8 | GO:0097623 | potassium ion export across plasma membrane(GO:0097623) |
| 1.5 | 4.5 | GO:0010814 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 1.4 | 11.5 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 1.4 | 6.9 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 1.2 | 3.6 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.9 | 9.5 | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 0.9 | 5.5 | GO:0021593 | rhombomere morphogenesis(GO:0021593) |
| 0.9 | 2.6 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) odontoblast differentiation(GO:0071895) regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 0.7 | 2.7 | GO:0051295 | establishment of meiotic spindle localization(GO:0051295) |
| 0.6 | 2.5 | GO:0060025 | regulation of synaptic activity(GO:0060025) |
| 0.6 | 1.8 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.6 | 9.8 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.5 | 6.3 | GO:1903504 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.4 | 0.8 | GO:0090135 | actin filament branching(GO:0090135) |
| 0.4 | 3.5 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.4 | 2.2 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.4 | 1.4 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 0.3 | 1.9 | GO:0042535 | positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.3 | 2.2 | GO:0072257 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.3 | 0.9 | GO:1901228 | positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 0.3 | 10.9 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.3 | 2.8 | GO:0003161 | cardiac conduction system development(GO:0003161) |
| 0.2 | 0.7 | GO:0045358 | negative regulation of interferon-beta biosynthetic process(GO:0045358) |
| 0.2 | 0.7 | GO:0010512 | negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) |
| 0.2 | 0.7 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.2 | 2.1 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.2 | 1.3 | GO:0045583 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.2 | 1.0 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.2 | 1.2 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.2 | 7.3 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.2 | 2.3 | GO:0050965 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.1 | 1.4 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 1.0 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 7.3 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.1 | 5.5 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.1 | 0.5 | GO:0002553 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) Golgi reassembly(GO:0090168) |
| 0.1 | 1.0 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.1 | 1.6 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.1 | 0.6 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 0.4 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 0.3 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 1.1 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 1.6 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 3.5 | GO:0007052 | mitotic spindle organization(GO:0007052) |
| 0.0 | 1.6 | GO:0006754 | ATP biosynthetic process(GO:0006754) |
| 0.0 | 1.4 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 5.2 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 1.2 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.7 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 6.9 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.9 | 4.5 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.5 | 2.7 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.3 | 3.5 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.3 | 1.3 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 0.2 | 6.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.2 | 2.6 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.2 | 1.8 | GO:0070938 | contractile ring(GO:0070938) |
| 0.2 | 1.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.2 | 3.5 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.2 | 1.5 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 1.6 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 1.0 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.1 | 4.2 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.1 | 2.8 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 4.7 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 0.8 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 0.7 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 1.0 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 2.8 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 3.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 3.5 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 1.3 | GO:0043034 | filamentous actin(GO:0031941) costamere(GO:0043034) |
| 0.0 | 3.5 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 0.5 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 8.0 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.8 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.8 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 2.2 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.7 | GO:0005902 | microvillus(GO:0005902) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 18.6 | GO:0070191 | methionine-R-sulfoxide reductase activity(GO:0070191) |
| 1.6 | 4.9 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.7 | 4.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.5 | 11.5 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.5 | 3.5 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.5 | 1.4 | GO:0018738 | S-formylglutathione hydrolase activity(GO:0018738) |
| 0.4 | 2.6 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.4 | 3.5 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.3 | 2.7 | GO:0043495 | protein anchor(GO:0043495) |
| 0.3 | 10.9 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.2 | 0.9 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.2 | 2.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.2 | 1.3 | GO:0002135 | CTP binding(GO:0002135) |
| 0.2 | 0.7 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.2 | 1.2 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.2 | 5.5 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.2 | 1.4 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 1.6 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.1 | 1.4 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 1.5 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 1.0 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 1.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 6.9 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 9.5 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 1.0 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 1.8 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 3.5 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 18.2 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 1.9 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 1.0 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 0.8 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 2.8 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 2.1 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 4.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.7 | GO:0048407 | platelet-derived growth factor receptor binding(GO:0005161) platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 12.3 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 1.6 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 1.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 3.6 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 3.6 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 1.6 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 2.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 11.0 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 1.7 | GO:0005178 | integrin binding(GO:0005178) |