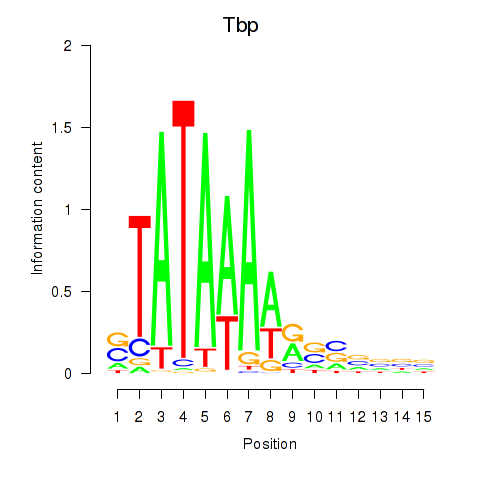
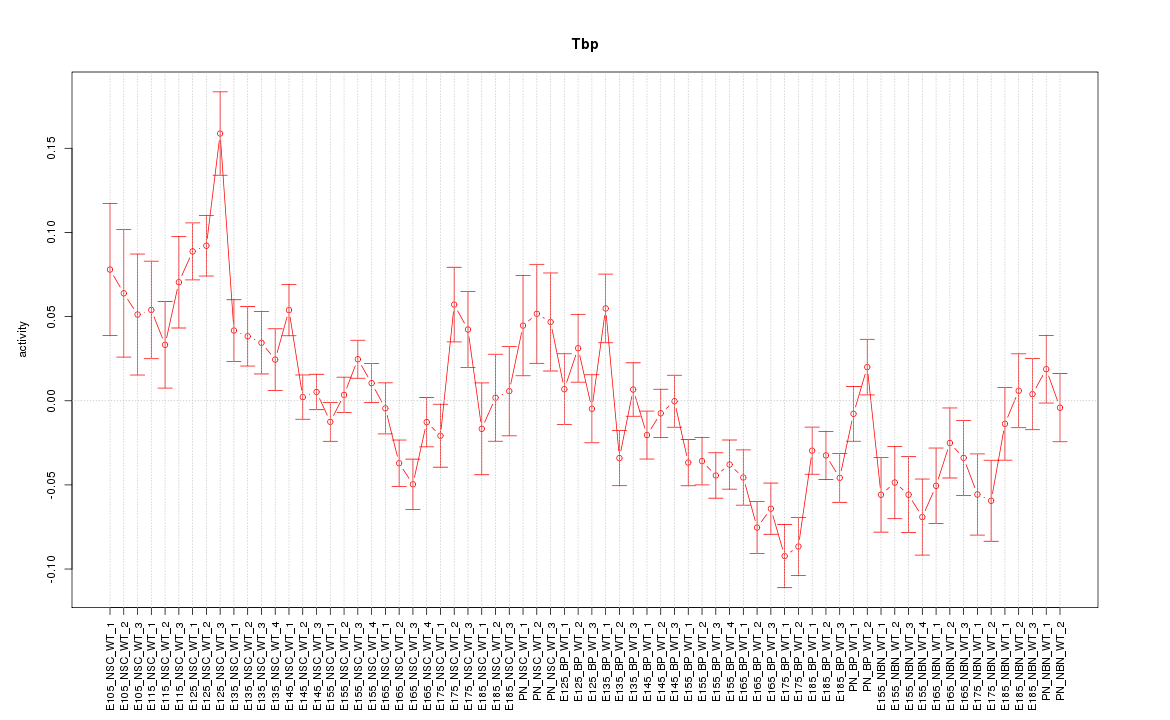
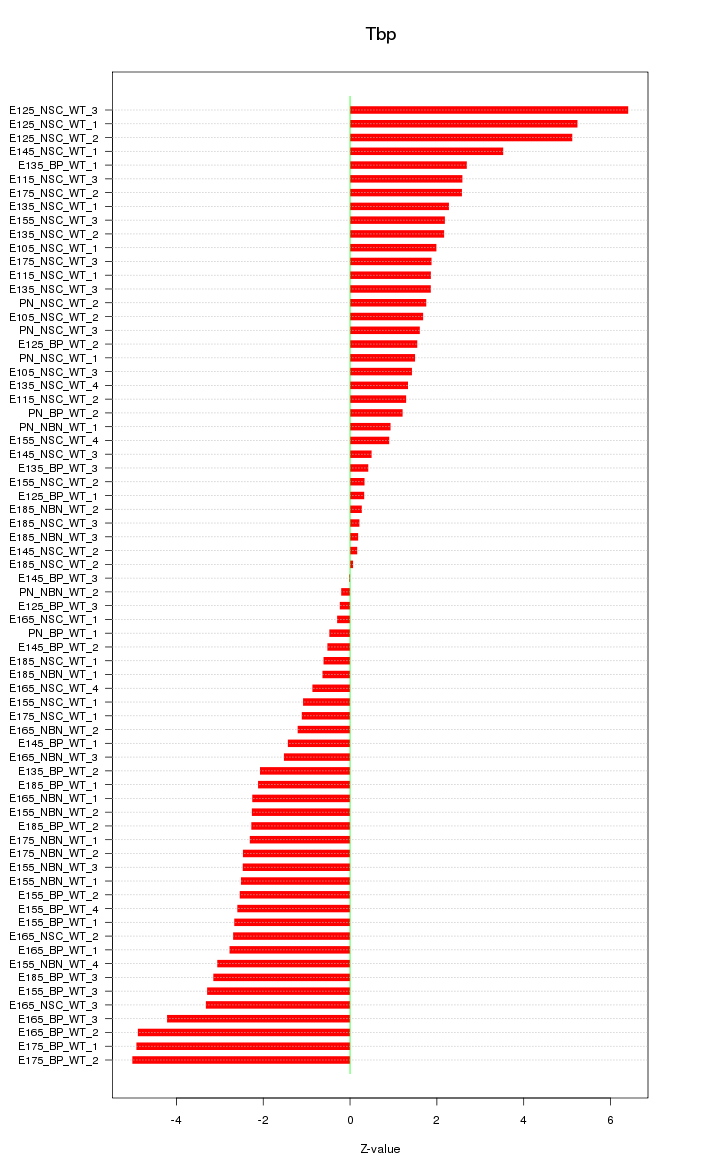
| 9.2 |
46.2 |
GO:0015671 |
oxygen transport(GO:0015671) |
| 8.9 |
26.8 |
GO:0021529 |
spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 8.9 |
26.6 |
GO:0003278 |
apoptotic process involved in heart morphogenesis(GO:0003278) |
| 4.5 |
22.3 |
GO:0010273 |
detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 3.9 |
15.7 |
GO:0045091 |
regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 3.4 |
13.4 |
GO:0035606 |
peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 3.3 |
13.2 |
GO:0042414 |
epinephrine metabolic process(GO:0042414) |
| 3.2 |
16.0 |
GO:1900147 |
Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 3.0 |
12.0 |
GO:0006407 |
rRNA export from nucleus(GO:0006407) |
| 2.9 |
11.8 |
GO:0032227 |
negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 2.8 |
14.1 |
GO:1903026 |
negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 2.7 |
10.9 |
GO:0042360 |
vitamin E metabolic process(GO:0042360) |
| 2.6 |
15.4 |
GO:0043415 |
positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 2.5 |
7.6 |
GO:0035519 |
protein K29-linked ubiquitination(GO:0035519) |
| 2.4 |
12.0 |
GO:0035262 |
gonad morphogenesis(GO:0035262) |
| 2.4 |
7.2 |
GO:2000852 |
regulation of corticosterone secretion(GO:2000852) |
| 2.4 |
31.2 |
GO:0042573 |
retinoic acid metabolic process(GO:0042573) positive regulation of collateral sprouting(GO:0048672) |
| 2.3 |
71.9 |
GO:0034723 |
DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 2.3 |
6.9 |
GO:0001951 |
intestinal D-glucose absorption(GO:0001951) terminal web assembly(GO:1902896) |
| 2.2 |
15.3 |
GO:0000320 |
re-entry into mitotic cell cycle(GO:0000320) |
| 2.2 |
6.5 |
GO:1900020 |
ovarian follicle rupture(GO:0001543) angiotensin mediated vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001998) regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) establishment of blood-nerve barrier(GO:0008065) regulation of activation of Janus kinase activity(GO:0010533) regulation of activation of JAK2 kinase activity(GO:0010534) regulation of amino acid import(GO:0010958) regulation of L-arginine import(GO:0010963) response to muscle activity involved in regulation of muscle adaptation(GO:0014873) regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) regulation of membrane hyperpolarization(GO:1902630) |
| 2.2 |
6.5 |
GO:0045472 |
response to ether(GO:0045472) |
| 2.0 |
5.9 |
GO:0070093 |
negative regulation of glucagon secretion(GO:0070093) |
| 2.0 |
5.9 |
GO:0006597 |
spermine biosynthetic process(GO:0006597) |
| 2.0 |
13.7 |
GO:0042590 |
antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 1.7 |
5.2 |
GO:0006059 |
hexitol metabolic process(GO:0006059) |
| 1.6 |
4.9 |
GO:0010899 |
regulation of phosphatidylcholine catabolic process(GO:0010899) |
| 1.6 |
3.3 |
GO:0036091 |
positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 1.6 |
9.4 |
GO:0030644 |
cellular chloride ion homeostasis(GO:0030644) |
| 1.6 |
4.7 |
GO:0060854 |
patterning of lymph vessels(GO:0060854) |
| 1.5 |
29.6 |
GO:0019430 |
removal of superoxide radicals(GO:0019430) |
| 1.4 |
8.5 |
GO:0043137 |
DNA replication, removal of RNA primer(GO:0043137) |
| 1.4 |
7.0 |
GO:0042535 |
positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 1.4 |
4.2 |
GO:1901896 |
positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 1.3 |
24.2 |
GO:0000920 |
cell separation after cytokinesis(GO:0000920) |
| 1.3 |
25.7 |
GO:0051044 |
positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 1.3 |
3.8 |
GO:0060558 |
regulation of calcidiol 1-monooxygenase activity(GO:0060558) |
| 1.2 |
3.7 |
GO:0042560 |
10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 1.2 |
8.5 |
GO:0016584 |
nucleosome positioning(GO:0016584) |
| 1.2 |
12.9 |
GO:0014807 |
regulation of somitogenesis(GO:0014807) |
| 1.1 |
8.0 |
GO:0046688 |
response to copper ion(GO:0046688) |
| 1.1 |
5.7 |
GO:0033087 |
negative regulation of immature T cell proliferation(GO:0033087) |
| 1.1 |
13.6 |
GO:0007076 |
mitotic chromosome condensation(GO:0007076) |
| 1.1 |
6.8 |
GO:2000210 |
positive regulation of anoikis(GO:2000210) |
| 1.1 |
2.2 |
GO:0019676 |
ammonia assimilation cycle(GO:0019676) |
| 1.1 |
6.7 |
GO:1904936 |
cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 1.1 |
15.2 |
GO:0038063 |
collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 1.1 |
9.6 |
GO:0042095 |
interferon-gamma biosynthetic process(GO:0042095) |
| 1.0 |
20.5 |
GO:0045662 |
negative regulation of myoblast differentiation(GO:0045662) |
| 1.0 |
3.8 |
GO:1902268 |
negative regulation of polyamine transmembrane transport(GO:1902268) |
| 1.0 |
2.9 |
GO:0015793 |
glycerol transport(GO:0015793) |
| 0.9 |
11.1 |
GO:0002227 |
innate immune response in mucosa(GO:0002227) |
| 0.9 |
3.5 |
GO:0015840 |
urea transport(GO:0015840) urea transmembrane transport(GO:0071918) |
| 0.9 |
3.5 |
GO:2000271 |
positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.8 |
14.0 |
GO:0021521 |
ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.8 |
12.4 |
GO:0035873 |
lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.8 |
10.7 |
GO:0001731 |
formation of translation preinitiation complex(GO:0001731) |
| 0.7 |
4.5 |
GO:0000050 |
urea cycle(GO:0000050) urea metabolic process(GO:0019627) |
| 0.7 |
2.2 |
GO:0050929 |
corticospinal neuron axon guidance through spinal cord(GO:0021972) positive regulation of negative chemotaxis(GO:0050924) induction of negative chemotaxis(GO:0050929) negative regulation of mononuclear cell migration(GO:0071676) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.7 |
5.0 |
GO:0042572 |
retinol metabolic process(GO:0042572) |
| 0.7 |
3.9 |
GO:1902998 |
macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of tau-protein kinase activity(GO:1902949) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.6 |
4.5 |
GO:0016560 |
protein import into peroxisome matrix, docking(GO:0016560) |
| 0.6 |
2.5 |
GO:0030299 |
intestinal cholesterol absorption(GO:0030299) lipid digestion(GO:0044241) |
| 0.6 |
4.2 |
GO:0001887 |
selenium compound metabolic process(GO:0001887) |
| 0.6 |
4.2 |
GO:1902416 |
positive regulation of mRNA binding(GO:1902416) positive regulation of RNA binding(GO:1905216) |
| 0.6 |
1.8 |
GO:0090285 |
negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.6 |
7.2 |
GO:0071803 |
positive regulation of podosome assembly(GO:0071803) |
| 0.5 |
35.3 |
GO:0006334 |
nucleosome assembly(GO:0006334) |
| 0.5 |
8.0 |
GO:0051451 |
myoblast migration(GO:0051451) |
| 0.5 |
7.9 |
GO:0060213 |
regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.4 |
6.5 |
GO:0050765 |
negative regulation of phagocytosis(GO:0050765) |
| 0.4 |
2.6 |
GO:1901029 |
negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.4 |
2.9 |
GO:0031642 |
negative regulation of myelination(GO:0031642) |
| 0.4 |
7.2 |
GO:0040034 |
regulation of development, heterochronic(GO:0040034) |
| 0.4 |
8.6 |
GO:0045672 |
positive regulation of osteoclast differentiation(GO:0045672) |
| 0.4 |
2.3 |
GO:0071638 |
cellular triglyceride homeostasis(GO:0035356) negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.4 |
6.7 |
GO:0051984 |
positive regulation of chromosome segregation(GO:0051984) |
| 0.3 |
1.6 |
GO:0010624 |
regulation of Schwann cell proliferation(GO:0010624) |
| 0.3 |
1.3 |
GO:0035878 |
nail development(GO:0035878) |
| 0.3 |
0.9 |
GO:1902310 |
positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.3 |
4.4 |
GO:0010640 |
regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) |
| 0.3 |
6.4 |
GO:0060216 |
definitive hemopoiesis(GO:0060216) |
| 0.2 |
5.4 |
GO:0071470 |
cellular response to osmotic stress(GO:0071470) |
| 0.2 |
16.4 |
GO:0030512 |
negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.2 |
6.0 |
GO:0071353 |
cellular response to interleukin-4(GO:0071353) |
| 0.2 |
3.8 |
GO:0034389 |
lipid particle organization(GO:0034389) |
| 0.2 |
14.6 |
GO:0002088 |
lens development in camera-type eye(GO:0002088) |
| 0.2 |
0.4 |
GO:1904139 |
microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.2 |
1.0 |
GO:0016127 |
cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.2 |
0.5 |
GO:0006556 |
S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.2 |
0.5 |
GO:1903225 |
regulation of endodermal cell fate specification(GO:0042663) negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.2 |
1.6 |
GO:0035970 |
peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.2 |
0.6 |
GO:0070213 |
protein auto-ADP-ribosylation(GO:0070213) |
| 0.1 |
6.5 |
GO:0033119 |
negative regulation of RNA splicing(GO:0033119) |
| 0.1 |
4.5 |
GO:0006730 |
one-carbon metabolic process(GO:0006730) |
| 0.1 |
1.3 |
GO:0010992 |
ubiquitin homeostasis(GO:0010992) |
| 0.1 |
3.6 |
GO:0034629 |
cellular protein complex localization(GO:0034629) |
| 0.1 |
0.4 |
GO:0001580 |
detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 |
1.0 |
GO:0036506 |
maintenance of unfolded protein(GO:0036506) protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.1 |
1.1 |
GO:0060837 |
blood vessel endothelial cell differentiation(GO:0060837) |
| 0.1 |
4.1 |
GO:0000387 |
spliceosomal snRNP assembly(GO:0000387) |
| 0.1 |
10.2 |
GO:0051225 |
spindle assembly(GO:0051225) |
| 0.1 |
5.6 |
GO:0022904 |
respiratory electron transport chain(GO:0022904) |
| 0.1 |
0.7 |
GO:0002002 |
regulation of angiotensin levels in blood(GO:0002002) |
| 0.1 |
7.4 |
GO:0010923 |
negative regulation of phosphatase activity(GO:0010923) |
| 0.1 |
2.7 |
GO:0000027 |
ribosomal large subunit assembly(GO:0000027) |
| 0.1 |
0.2 |
GO:0002408 |
myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.1 |
1.9 |
GO:1903427 |
negative regulation of reactive oxygen species biosynthetic process(GO:1903427) |
| 0.1 |
1.0 |
GO:0007175 |
negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 |
0.6 |
GO:0000712 |
resolution of meiotic recombination intermediates(GO:0000712) |
| 0.1 |
0.8 |
GO:2000051 |
negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.1 |
0.6 |
GO:0007000 |
nucleolus organization(GO:0007000) |
| 0.1 |
3.9 |
GO:0031338 |
regulation of vesicle fusion(GO:0031338) |
| 0.1 |
1.1 |
GO:0097286 |
iron ion import(GO:0097286) |
| 0.1 |
9.7 |
GO:0030217 |
T cell differentiation(GO:0030217) |
| 0.1 |
2.9 |
GO:0098780 |
mitophagy in response to mitochondrial depolarization(GO:0098779) response to mitochondrial depolarisation(GO:0098780) |
| 0.1 |
3.0 |
GO:0061515 |
myeloid cell development(GO:0061515) |
| 0.1 |
2.6 |
GO:0006418 |
tRNA aminoacylation for protein translation(GO:0006418) |
| 0.1 |
0.2 |
GO:0030277 |
maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 |
1.6 |
GO:0007608 |
sensory perception of smell(GO:0007608) |
| 0.0 |
13.5 |
GO:0007067 |
mitotic nuclear division(GO:0007067) |
| 0.0 |
3.1 |
GO:1903955 |
positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 |
0.7 |
GO:0043171 |
peptide catabolic process(GO:0043171) |
| 0.0 |
0.9 |
GO:0015985 |
energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 |
3.5 |
GO:0051028 |
mRNA transport(GO:0051028) |
| 0.0 |
4.1 |
GO:0008033 |
tRNA processing(GO:0008033) |
| 0.0 |
0.9 |
GO:0095500 |
acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.0 |
0.5 |
GO:0071480 |
cellular response to gamma radiation(GO:0071480) |
| 0.0 |
1.4 |
GO:0030500 |
regulation of bone mineralization(GO:0030500) |
| 0.0 |
3.5 |
GO:0007519 |
skeletal muscle tissue development(GO:0007519) |
| 0.0 |
4.1 |
GO:0042254 |
ribosome biogenesis(GO:0042254) |
| 0.0 |
0.8 |
GO:0031397 |
negative regulation of protein ubiquitination(GO:0031397) |
| 0.0 |
0.5 |
GO:0006414 |
translational elongation(GO:0006414) |
| 0.0 |
0.5 |
GO:0006352 |
DNA-templated transcription, initiation(GO:0006352) |
| 0.0 |
0.2 |
GO:0035518 |
histone H2A monoubiquitination(GO:0035518) |