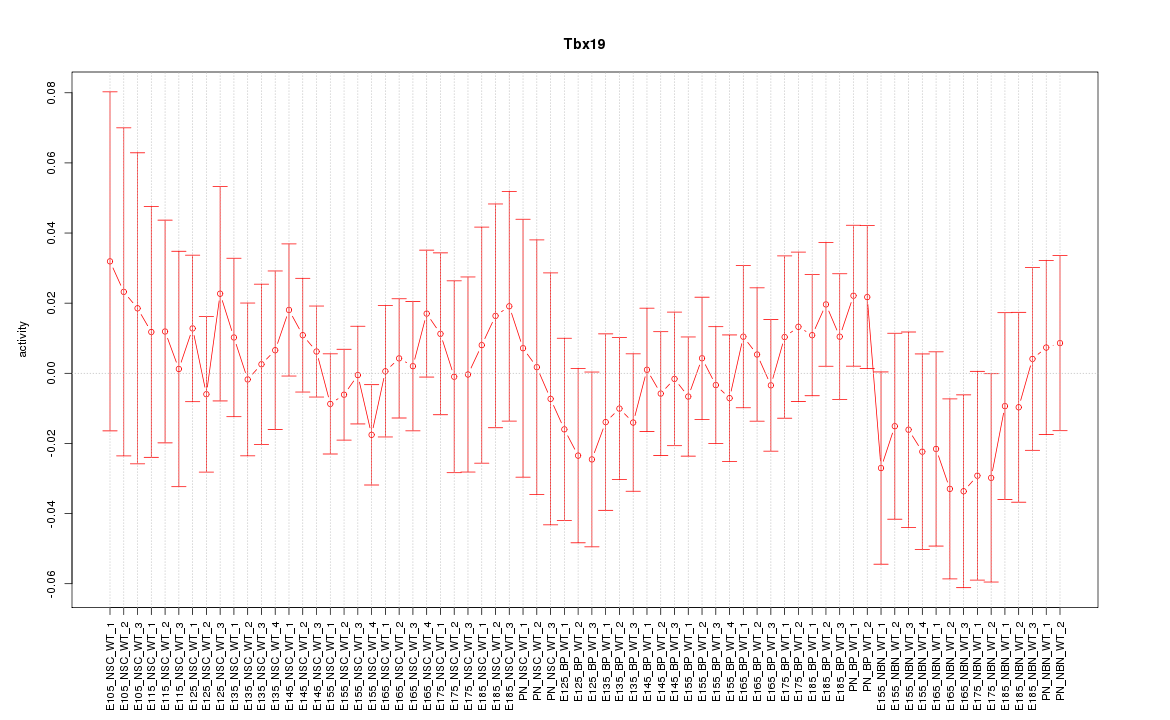
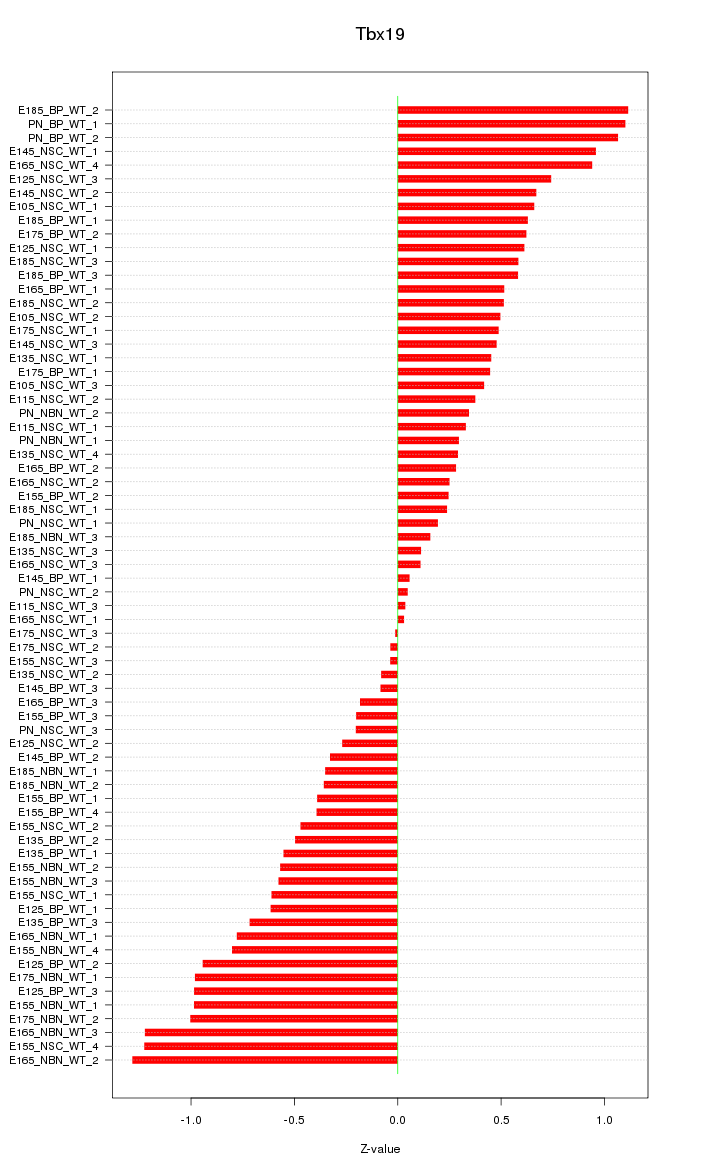
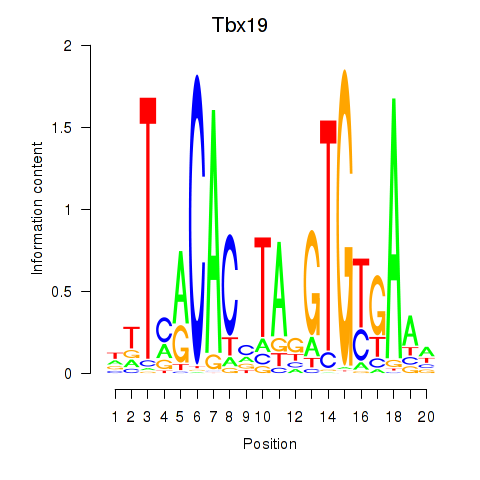
Motif ID: Tbx19
Z-value: 0.605
Transcription factors associated with Tbx19:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Tbx19 | ENSMUSG00000026572.5 | Tbx19 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0002538 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) olefin metabolic process(GO:1900673) |
| 0.7 | 2.7 | GO:1902990 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.5 | 2.5 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.4 | 2.5 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.4 | 1.1 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.4 | 1.1 | GO:1901228 | regulation of osteoclast proliferation(GO:0090289) negative regulation of bone mineralization involved in bone maturation(GO:1900158) positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 0.3 | 1.4 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.3 | 1.3 | GO:0071033 | nuclear retention of pre-mRNA at the site of transcription(GO:0071033) CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.3 | 2.5 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.3 | 1.5 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.3 | 1.0 | GO:0051462 | cortisol secretion(GO:0043400) regulation of cortisol secretion(GO:0051462) |
| 0.2 | 1.4 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.2 | 1.1 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.2 | 0.6 | GO:0097278 | complement-dependent cytotoxicity(GO:0097278) |
| 0.2 | 1.2 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.2 | 2.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.2 | 0.5 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.2 | 1.0 | GO:0098734 | protein depalmitoylation(GO:0002084) positive regulation of pinocytosis(GO:0048549) macromolecule depalmitoylation(GO:0098734) |
| 0.2 | 2.1 | GO:0072010 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.1 | 0.4 | GO:0031590 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.1 | 1.0 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.1 | 0.4 | GO:0045819 | positive regulation of glycogen catabolic process(GO:0045819) |
| 0.1 | 1.3 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.1 | 0.8 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 0.3 | GO:0010958 | regulation of amino acid import(GO:0010958) regulation of L-arginine import(GO:0010963) |
| 0.1 | 3.2 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.1 | 1.4 | GO:0042640 | anagen(GO:0042640) |
| 0.1 | 0.2 | GO:0045014 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) detection of glucose(GO:0051594) |
| 0.1 | 0.4 | GO:0032627 | interleukin-23 production(GO:0032627) regulation of interleukin-23 production(GO:0032667) |
| 0.1 | 0.5 | GO:0097460 | ferrous iron import into cell(GO:0097460) |
| 0.1 | 0.4 | GO:0032532 | regulation of microvillus length(GO:0032532) |
| 0.1 | 0.4 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.1 | 1.0 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 0.2 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.1 | 0.3 | GO:0002677 | negative regulation of chronic inflammatory response(GO:0002677) |
| 0.1 | 1.4 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 0.7 | GO:1990173 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.1 | 3.8 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 2.5 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.6 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.3 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.3 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.0 | 0.5 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.0 | 0.4 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.2 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.3 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.5 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.0 | 0.4 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 2.6 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.9 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.0 | 0.6 | GO:0032543 | mitochondrial translation(GO:0032543) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.8 | 4.7 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.3 | 2.6 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.2 | 1.3 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 0.6 | GO:0033093 | multivesicular body membrane(GO:0032585) Weibel-Palade body(GO:0033093) |
| 0.1 | 1.3 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 1.4 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 0.6 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 0.5 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 0.7 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 2.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.1 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.4 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.2 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 2.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.5 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.6 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 5.6 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.3 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.7 | GO:0031526 | brush border membrane(GO:0031526) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) dinucleotide insertion or deletion binding(GO:0032139) |
| 0.4 | 2.5 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.3 | 2.0 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.3 | 1.5 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.3 | 1.4 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.2 | 0.7 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.2 | 2.9 | GO:0008430 | selenium binding(GO:0008430) |
| 0.2 | 0.6 | GO:0034618 | arginine binding(GO:0034618) |
| 0.2 | 1.1 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.2 | 0.5 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.1 | 3.8 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 1.1 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 0.1 | 1.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 1.3 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.1 | 2.5 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 0.4 | GO:0000402 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.1 | 1.1 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.1 | 1.2 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 1.0 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 1.8 | GO:0043747 | protein-N-terminal asparagine amidohydrolase activity(GO:0008418) UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase activity(GO:0008759) iprodione amidohydrolase activity(GO:0018748) (3,5-dichlorophenylurea)acetate amidohydrolase activity(GO:0018749) 4'-(2-hydroxyisopropyl)phenylurea amidohydrolase activity(GO:0034571) didemethylisoproturon amidohydrolase activity(GO:0034573) N-isopropylacetanilide amidohydrolase activity(GO:0034576) N-cyclohexylformamide amidohydrolase activity(GO:0034781) isonicotinic acid hydrazide hydrolase activity(GO:0034876) cis-aconitamide amidase activity(GO:0034882) gamma-N-formylaminovinylacetate hydrolase activity(GO:0034885) N2-acetyl-L-lysine deacetylase activity(GO:0043747) O-succinylbenzoate synthase activity(GO:0043748) indoleacetamide hydrolase activity(GO:0043864) N-acetylcitrulline deacetylase activity(GO:0043909) N-acetylgalactosamine-6-phosphate deacetylase activity(GO:0047419) diacetylchitobiose deacetylase activity(GO:0052773) chitooligosaccharide deacetylase activity(GO:0052790) |
| 0.1 | 0.2 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 0.5 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.1 | 0.3 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 2.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.4 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.1 | 0.3 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.4 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.3 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.7 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 2.5 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.0 | 0.6 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.3 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 0.3 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.1 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) |
| 0.0 | 2.9 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 2.9 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 1.0 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 2.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |