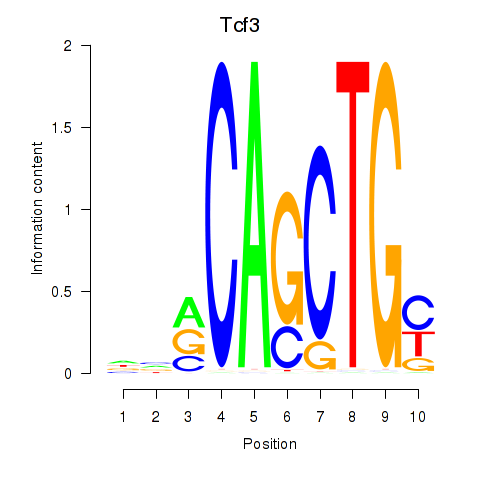
| 2.4 |
7.1 |
GO:0006601 |
creatine biosynthetic process(GO:0006601) |
| 2.2 |
8.7 |
GO:0060032 |
notochord regression(GO:0060032) |
| 1.9 |
7.7 |
GO:0046836 |
glycolipid transport(GO:0046836) |
| 1.9 |
5.6 |
GO:0000087 |
mitotic M phase(GO:0000087) |
| 1.5 |
1.5 |
GO:0042321 |
negative regulation of circadian sleep/wake cycle, sleep(GO:0042321) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 1.5 |
4.6 |
GO:0032650 |
regulation of interleukin-1 alpha production(GO:0032650) positive regulation of interleukin-1 alpha production(GO:0032730) cardiac cell fate determination(GO:0060913) |
| 1.5 |
4.4 |
GO:0010814 |
substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 1.4 |
7.1 |
GO:0014719 |
skeletal muscle satellite cell activation(GO:0014719) |
| 1.2 |
3.5 |
GO:1904879 |
positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 1.1 |
5.7 |
GO:0007386 |
compartment pattern specification(GO:0007386) |
| 1.1 |
6.7 |
GO:0006287 |
base-excision repair, gap-filling(GO:0006287) |
| 1.1 |
13.0 |
GO:0060272 |
embryonic skeletal joint morphogenesis(GO:0060272) |
| 1.0 |
2.1 |
GO:0072197 |
ureter morphogenesis(GO:0072197) |
| 1.0 |
11.4 |
GO:0061302 |
smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.9 |
4.7 |
GO:0048133 |
germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.9 |
3.6 |
GO:0010288 |
response to lead ion(GO:0010288) |
| 0.9 |
8.0 |
GO:0045603 |
positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.9 |
2.6 |
GO:0006546 |
glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.9 |
2.6 |
GO:1903998 |
regulation of eating behavior(GO:1903998) |
| 0.8 |
7.6 |
GO:0042487 |
regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.8 |
2.4 |
GO:0042939 |
renal sodium ion transport(GO:0003096) glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.8 |
3.8 |
GO:0046598 |
positive regulation of viral entry into host cell(GO:0046598) |
| 0.8 |
3.0 |
GO:0032439 |
endosome localization(GO:0032439) |
| 0.8 |
2.3 |
GO:0003100 |
regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) |
| 0.7 |
3.7 |
GO:0006564 |
L-serine biosynthetic process(GO:0006564) |
| 0.7 |
3.7 |
GO:0010668 |
ectodermal cell differentiation(GO:0010668) |
| 0.7 |
2.9 |
GO:0016340 |
calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.7 |
5.0 |
GO:2000491 |
positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.7 |
7.7 |
GO:0042473 |
outer ear morphogenesis(GO:0042473) |
| 0.7 |
3.3 |
GO:0046864 |
retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) alveolar primary septum development(GO:0061143) |
| 0.7 |
4.6 |
GO:0021984 |
adenohypophysis development(GO:0021984) |
| 0.7 |
2.0 |
GO:0014835 |
myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.7 |
3.3 |
GO:1900085 |
negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) |
| 0.6 |
3.2 |
GO:0071205 |
clustering of voltage-gated potassium channels(GO:0045163) protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.6 |
2.5 |
GO:2001271 |
negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.6 |
2.5 |
GO:1990414 |
replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.6 |
4.3 |
GO:0051461 |
positive regulation of heat generation(GO:0031652) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.6 |
2.4 |
GO:0060715 |
syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.6 |
2.3 |
GO:0042905 |
9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.6 |
5.7 |
GO:0001977 |
renal system process involved in regulation of blood volume(GO:0001977) |
| 0.6 |
5.5 |
GO:0030322 |
stabilization of membrane potential(GO:0030322) |
| 0.6 |
11.6 |
GO:0032967 |
positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.5 |
2.1 |
GO:0045578 |
negative regulation of B cell differentiation(GO:0045578) |
| 0.5 |
2.1 |
GO:1900220 |
semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.5 |
6.2 |
GO:0014842 |
skeletal muscle satellite cell proliferation(GO:0014841) regulation of skeletal muscle satellite cell proliferation(GO:0014842) |
| 0.5 |
1.5 |
GO:0035582 |
sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.5 |
4.1 |
GO:0035735 |
intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.5 |
3.5 |
GO:0070458 |
detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.5 |
1.5 |
GO:0015772 |
disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.5 |
1.5 |
GO:0030167 |
proteoglycan catabolic process(GO:0030167) |
| 0.5 |
6.0 |
GO:0060159 |
regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.5 |
2.0 |
GO:0048539 |
bone marrow development(GO:0048539) |
| 0.5 |
1.4 |
GO:1903048 |
regulation of acetylcholine-gated cation channel activity(GO:1903048) |
| 0.5 |
2.8 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.5 |
1.4 |
GO:0021577 |
hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.5 |
2.7 |
GO:0009449 |
gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.4 |
1.3 |
GO:0044333 |
Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.4 |
1.3 |
GO:2000564 |
regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) CD8-positive, alpha-beta T cell proliferation(GO:0035740) negative regulation of regulatory T cell differentiation(GO:0045590) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.4 |
0.9 |
GO:2000507 |
positive regulation of energy homeostasis(GO:2000507) |
| 0.4 |
6.8 |
GO:0040034 |
regulation of development, heterochronic(GO:0040034) |
| 0.4 |
3.0 |
GO:0002315 |
marginal zone B cell differentiation(GO:0002315) |
| 0.4 |
1.3 |
GO:0034162 |
toll-like receptor 9 signaling pathway(GO:0034162) |
| 0.4 |
2.9 |
GO:0060923 |
cardiac muscle cell fate commitment(GO:0060923) |
| 0.4 |
2.1 |
GO:0071500 |
cellular response to nitrosative stress(GO:0071500) |
| 0.4 |
1.2 |
GO:2001054 |
negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.4 |
2.0 |
GO:0014016 |
neuroblast differentiation(GO:0014016) response to folic acid(GO:0051593) |
| 0.4 |
1.2 |
GO:2000409 |
positive regulation of T cell extravasation(GO:2000409) |
| 0.4 |
6.8 |
GO:0051988 |
regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.4 |
1.1 |
GO:0002901 |
mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.4 |
5.5 |
GO:0030903 |
notochord development(GO:0030903) |
| 0.4 |
1.1 |
GO:0035574 |
histone H4-K20 demethylation(GO:0035574) |
| 0.4 |
2.1 |
GO:0014816 |
skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.3 |
1.7 |
GO:0001561 |
fatty acid alpha-oxidation(GO:0001561) |
| 0.3 |
10.3 |
GO:0045746 |
negative regulation of Notch signaling pathway(GO:0045746) |
| 0.3 |
1.3 |
GO:0006933 |
negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.3 |
2.0 |
GO:0021593 |
rhombomere morphogenesis(GO:0021593) |
| 0.3 |
1.0 |
GO:0030202 |
heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) heparin metabolic process(GO:0030202) |
| 0.3 |
8.5 |
GO:0045747 |
positive regulation of Notch signaling pathway(GO:0045747) |
| 0.3 |
1.2 |
GO:0033580 |
protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.3 |
0.9 |
GO:0045819 |
positive regulation of glycogen catabolic process(GO:0045819) |
| 0.3 |
0.9 |
GO:1902202 |
regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.3 |
1.4 |
GO:2000680 |
regulation of rubidium ion transport(GO:2000680) |
| 0.3 |
3.4 |
GO:0097067 |
cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.3 |
2.3 |
GO:1900262 |
regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.3 |
0.8 |
GO:0030327 |
prenylated protein catabolic process(GO:0030327) |
| 0.3 |
2.0 |
GO:0002693 |
positive regulation of cellular extravasation(GO:0002693) |
| 0.3 |
0.8 |
GO:0043654 |
recognition of apoptotic cell(GO:0043654) |
| 0.3 |
1.1 |
GO:0072235 |
distal convoluted tubule development(GO:0072025) DCT cell differentiation(GO:0072069) metanephric distal convoluted tubule development(GO:0072221) metanephric distal tubule development(GO:0072235) metanephric DCT cell differentiation(GO:0072240) |
| 0.3 |
1.0 |
GO:0002002 |
regulation of angiotensin levels in blood(GO:0002002) |
| 0.3 |
5.0 |
GO:0034142 |
toll-like receptor 4 signaling pathway(GO:0034142) |
| 0.3 |
0.8 |
GO:0050859 |
negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.3 |
4.1 |
GO:0014823 |
response to activity(GO:0014823) |
| 0.3 |
3.3 |
GO:0071803 |
positive regulation of podosome assembly(GO:0071803) |
| 0.2 |
1.9 |
GO:0038166 |
angiotensin-activated signaling pathway(GO:0038166) |
| 0.2 |
2.1 |
GO:0021678 |
third ventricle development(GO:0021678) |
| 0.2 |
0.7 |
GO:2000832 |
protein-chromophore linkage(GO:0018298) negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.2 |
2.0 |
GO:0006450 |
regulation of translational fidelity(GO:0006450) |
| 0.2 |
2.6 |
GO:0032463 |
negative regulation of protein homooligomerization(GO:0032463) |
| 0.2 |
1.0 |
GO:2000623 |
regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.2 |
1.2 |
GO:0019336 |
phenol-containing compound catabolic process(GO:0019336) |
| 0.2 |
0.6 |
GO:0006436 |
tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.2 |
2.0 |
GO:0006560 |
proline metabolic process(GO:0006560) |
| 0.2 |
1.6 |
GO:0031848 |
protection from non-homologous end joining at telomere(GO:0031848) |
| 0.2 |
1.7 |
GO:0042534 |
tumor necrosis factor biosynthetic process(GO:0042533) regulation of tumor necrosis factor biosynthetic process(GO:0042534) |
| 0.2 |
1.1 |
GO:0044336 |
canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.2 |
0.9 |
GO:0090666 |
telomere assembly(GO:0032202) scaRNA localization to Cajal body(GO:0090666) |
| 0.2 |
1.3 |
GO:0051004 |
regulation of lipoprotein lipase activity(GO:0051004) |
| 0.2 |
2.0 |
GO:0009650 |
UV protection(GO:0009650) |
| 0.2 |
0.9 |
GO:0034115 |
negative regulation of heterotypic cell-cell adhesion(GO:0034115) cell-cell adhesion involved in gastrulation(GO:0070586) regulation of cell-cell adhesion involved in gastrulation(GO:0070587) negative regulation of protein kinase C signaling(GO:0090038) |
| 0.2 |
1.4 |
GO:0015868 |
purine ribonucleotide transport(GO:0015868) |
| 0.2 |
0.4 |
GO:0003199 |
endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.2 |
0.5 |
GO:1901675 |
negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.2 |
2.3 |
GO:0032957 |
negative regulation of platelet-derived growth factor receptor signaling pathway(GO:0010642) inositol trisphosphate metabolic process(GO:0032957) |
| 0.2 |
1.0 |
GO:2000210 |
positive regulation of anoikis(GO:2000210) |
| 0.2 |
0.5 |
GO:0045014 |
ectoderm formation(GO:0001705) detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) detection of glucose(GO:0051594) |
| 0.1 |
0.9 |
GO:0032534 |
regulation of microvillus assembly(GO:0032534) |
| 0.1 |
1.9 |
GO:2000353 |
positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.1 |
0.3 |
GO:0001803 |
type III hypersensitivity(GO:0001802) regulation of type III hypersensitivity(GO:0001803) positive regulation of type III hypersensitivity(GO:0001805) |
| 0.1 |
1.8 |
GO:0048642 |
negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.1 |
1.2 |
GO:0016127 |
cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 |
1.6 |
GO:0006610 |
ribosomal protein import into nucleus(GO:0006610) |
| 0.1 |
1.0 |
GO:0051418 |
interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.1 |
1.0 |
GO:0010571 |
positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.1 |
0.7 |
GO:0038018 |
Wnt receptor catabolic process(GO:0038018) |
| 0.1 |
1.1 |
GO:0043586 |
tongue development(GO:0043586) |
| 0.1 |
1.6 |
GO:0061299 |
retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.1 |
0.7 |
GO:0009052 |
pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.1 |
0.6 |
GO:0038032 |
termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.1 |
1.1 |
GO:0090050 |
peptidyl-lysine deacetylation(GO:0034983) regulation of skeletal muscle fiber development(GO:0048742) positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 |
7.7 |
GO:0051489 |
regulation of filopodium assembly(GO:0051489) |
| 0.1 |
2.5 |
GO:0010569 |
regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.1 |
3.3 |
GO:0050873 |
brown fat cell differentiation(GO:0050873) |
| 0.1 |
0.5 |
GO:1901341 |
activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 |
0.6 |
GO:1904690 |
adenosine to inosine editing(GO:0006382) regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.1 |
2.2 |
GO:1902857 |
positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.1 |
0.5 |
GO:0034351 |
regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.1 |
1.8 |
GO:2000001 |
regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 |
0.5 |
GO:0065001 |
negative regulation of histone H3-K36 methylation(GO:0000415) specification of axis polarity(GO:0065001) |
| 0.1 |
0.6 |
GO:0051895 |
negative regulation of focal adhesion assembly(GO:0051895) negative regulation of adherens junction organization(GO:1903392) |
| 0.1 |
1.5 |
GO:0060736 |
prostate gland growth(GO:0060736) |
| 0.1 |
0.3 |
GO:0006706 |
steroid catabolic process(GO:0006706) |
| 0.1 |
0.7 |
GO:0090527 |
actin filament reorganization(GO:0090527) |
| 0.1 |
1.2 |
GO:0010748 |
negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.1 |
5.5 |
GO:0048384 |
retinoic acid receptor signaling pathway(GO:0048384) |
| 0.1 |
0.6 |
GO:0006172 |
ADP biosynthetic process(GO:0006172) |
| 0.1 |
0.7 |
GO:2000124 |
regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 |
0.4 |
GO:0061009 |
common bile duct development(GO:0061009) |
| 0.1 |
1.6 |
GO:0032292 |
myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.1 |
1.1 |
GO:0090557 |
establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 |
1.0 |
GO:1901409 |
regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 |
0.8 |
GO:0000185 |
activation of MAPKKK activity(GO:0000185) |
| 0.1 |
4.1 |
GO:0002011 |
morphogenesis of an epithelial sheet(GO:0002011) |
| 0.1 |
1.1 |
GO:0032780 |
negative regulation of ATPase activity(GO:0032780) |
| 0.1 |
5.7 |
GO:0070373 |
negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.1 |
0.4 |
GO:0006867 |
asparagine transport(GO:0006867) glutamine transport(GO:0006868) |
| 0.1 |
1.4 |
GO:0045662 |
negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 |
1.2 |
GO:0021924 |
cell proliferation in external granule layer(GO:0021924) cerebellar granule cell precursor proliferation(GO:0021930) |
| 0.1 |
1.8 |
GO:0031954 |
positive regulation of protein autophosphorylation(GO:0031954) |
| 0.1 |
1.4 |
GO:0032288 |
myelin assembly(GO:0032288) |
| 0.1 |
2.0 |
GO:0034629 |
cellular protein complex localization(GO:0034629) |
| 0.1 |
1.5 |
GO:0042104 |
positive regulation of activated T cell proliferation(GO:0042104) |
| 0.1 |
2.2 |
GO:0032008 |
positive regulation of TOR signaling(GO:0032008) |
| 0.1 |
1.4 |
GO:0021511 |
spinal cord patterning(GO:0021511) |
| 0.1 |
3.0 |
GO:0060612 |
adipose tissue development(GO:0060612) |
| 0.1 |
0.4 |
GO:2000726 |
negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) negative regulation of cardiocyte differentiation(GO:1905208) negative regulation of cardiac muscle cell differentiation(GO:2000726) |
| 0.1 |
1.0 |
GO:0046597 |
negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 |
0.1 |
GO:2000402 |
negative regulation of lymphocyte migration(GO:2000402) |
| 0.1 |
2.0 |
GO:0010842 |
retina layer formation(GO:0010842) |
| 0.1 |
3.3 |
GO:0030835 |
negative regulation of actin filament depolymerization(GO:0030835) |
| 0.1 |
2.5 |
GO:0006730 |
one-carbon metabolic process(GO:0006730) |
| 0.1 |
0.6 |
GO:0045103 |
intermediate filament-based process(GO:0045103) |
| 0.1 |
0.6 |
GO:0010457 |
centriole-centriole cohesion(GO:0010457) |
| 0.1 |
2.2 |
GO:0035329 |
hippo signaling(GO:0035329) |
| 0.1 |
0.3 |
GO:0050853 |
B cell receptor signaling pathway(GO:0050853) |
| 0.1 |
0.5 |
GO:0036506 |
maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.1 |
1.0 |
GO:0032897 |
negative regulation of viral transcription(GO:0032897) |
| 0.1 |
0.3 |
GO:0061032 |
visceral serous pericardium development(GO:0061032) |
| 0.1 |
0.7 |
GO:1902287 |
semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.1 |
2.2 |
GO:0006940 |
regulation of smooth muscle contraction(GO:0006940) |
| 0.1 |
1.4 |
GO:0060706 |
cell differentiation involved in embryonic placenta development(GO:0060706) |
| 0.1 |
0.5 |
GO:0035066 |
positive regulation of histone acetylation(GO:0035066) |
| 0.1 |
0.3 |
GO:0051152 |
positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.1 |
1.6 |
GO:0010633 |
negative regulation of epithelial cell migration(GO:0010633) |
| 0.0 |
0.0 |
GO:0015938 |
coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) |
| 0.0 |
0.7 |
GO:0006517 |
protein deglycosylation(GO:0006517) |
| 0.0 |
1.3 |
GO:2001222 |
regulation of neuron migration(GO:2001222) |
| 0.0 |
1.4 |
GO:0001658 |
branching involved in ureteric bud morphogenesis(GO:0001658) |
| 0.0 |
1.5 |
GO:0017157 |
regulation of exocytosis(GO:0017157) |
| 0.0 |
1.3 |
GO:0000154 |
rRNA modification(GO:0000154) |
| 0.0 |
0.4 |
GO:0043619 |
regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 |
0.6 |
GO:0006910 |
phagocytosis, recognition(GO:0006910) |
| 0.0 |
0.1 |
GO:0008050 |
female courtship behavior(GO:0008050) |
| 0.0 |
0.3 |
GO:0031914 |
negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 |
0.3 |
GO:0032790 |
ribosome disassembly(GO:0032790) |
| 0.0 |
0.2 |
GO:0035988 |
chondrocyte proliferation(GO:0035988) |
| 0.0 |
0.7 |
GO:0010831 |
positive regulation of myotube differentiation(GO:0010831) |
| 0.0 |
0.2 |
GO:0017156 |
calcium ion regulated exocytosis(GO:0017156) |
| 0.0 |
0.2 |
GO:0032911 |
neurotrophin production(GO:0032898) nerve growth factor production(GO:0032902) negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.0 |
0.3 |
GO:0009109 |
coenzyme catabolic process(GO:0009109) |
| 0.0 |
0.1 |
GO:0032512 |
regulation of protein phosphatase type 2B activity(GO:0032512) negative regulation of protein phosphatase type 2B activity(GO:0032513) |
| 0.0 |
0.7 |
GO:0006767 |
water-soluble vitamin metabolic process(GO:0006767) |
| 0.0 |
0.1 |
GO:0060011 |
Sertoli cell proliferation(GO:0060011) |
| 0.0 |
1.0 |
GO:0003333 |
amino acid transmembrane transport(GO:0003333) |
| 0.0 |
0.6 |
GO:0044804 |
nucleophagy(GO:0044804) |
| 0.0 |
1.1 |
GO:0046329 |
negative regulation of JNK cascade(GO:0046329) |
| 0.0 |
0.7 |
GO:0070534 |
protein K63-linked ubiquitination(GO:0070534) |
| 0.0 |
0.1 |
GO:0060481 |
lobar bronchus epithelium development(GO:0060481) |
| 0.0 |
0.4 |
GO:0048280 |
vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 |
0.6 |
GO:0070208 |
protein heterotrimerization(GO:0070208) |
| 0.0 |
2.8 |
GO:0007219 |
Notch signaling pathway(GO:0007219) |
| 0.0 |
0.2 |
GO:0006729 |
tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 |
1.6 |
GO:0070936 |
protein K48-linked ubiquitination(GO:0070936) |
| 0.0 |
1.5 |
GO:0030183 |
B cell differentiation(GO:0030183) |
| 0.0 |
0.4 |
GO:0016556 |
mRNA modification(GO:0016556) |
| 0.0 |
0.6 |
GO:0045600 |
positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 |
0.5 |
GO:0043242 |
negative regulation of microtubule depolymerization(GO:0007026) negative regulation of protein complex disassembly(GO:0043242) negative regulation of protein depolymerization(GO:1901880) |
| 0.0 |
0.5 |
GO:0000470 |
maturation of LSU-rRNA(GO:0000470) |
| 0.0 |
0.7 |
GO:0060325 |
face morphogenesis(GO:0060325) |
| 0.0 |
0.9 |
GO:0048663 |
neuron fate commitment(GO:0048663) |
| 0.0 |
0.3 |
GO:0046855 |
phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 |
0.3 |
GO:0015693 |
magnesium ion transport(GO:0015693) |
| 0.0 |
0.4 |
GO:0000132 |
establishment of mitotic spindle orientation(GO:0000132) |
| 0.0 |
2.2 |
GO:0019827 |
stem cell population maintenance(GO:0019827) maintenance of cell number(GO:0098727) |
| 0.0 |
0.9 |
GO:0061077 |
chaperone-mediated protein folding(GO:0061077) |
| 0.0 |
0.3 |
GO:0046854 |
phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 |
0.1 |
GO:0048739 |
cardiac muscle fiber development(GO:0048739) |
| 0.0 |
0.4 |
GO:0006368 |
transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 |
1.1 |
GO:0051297 |
centrosome organization(GO:0051297) |
| 0.0 |
0.6 |
GO:0016575 |
histone deacetylation(GO:0016575) |
| 0.0 |
0.3 |
GO:0007214 |
gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 |
0.1 |
GO:0030263 |
apoptotic chromosome condensation(GO:0030263) |
| 0.0 |
0.1 |
GO:0016338 |
calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |