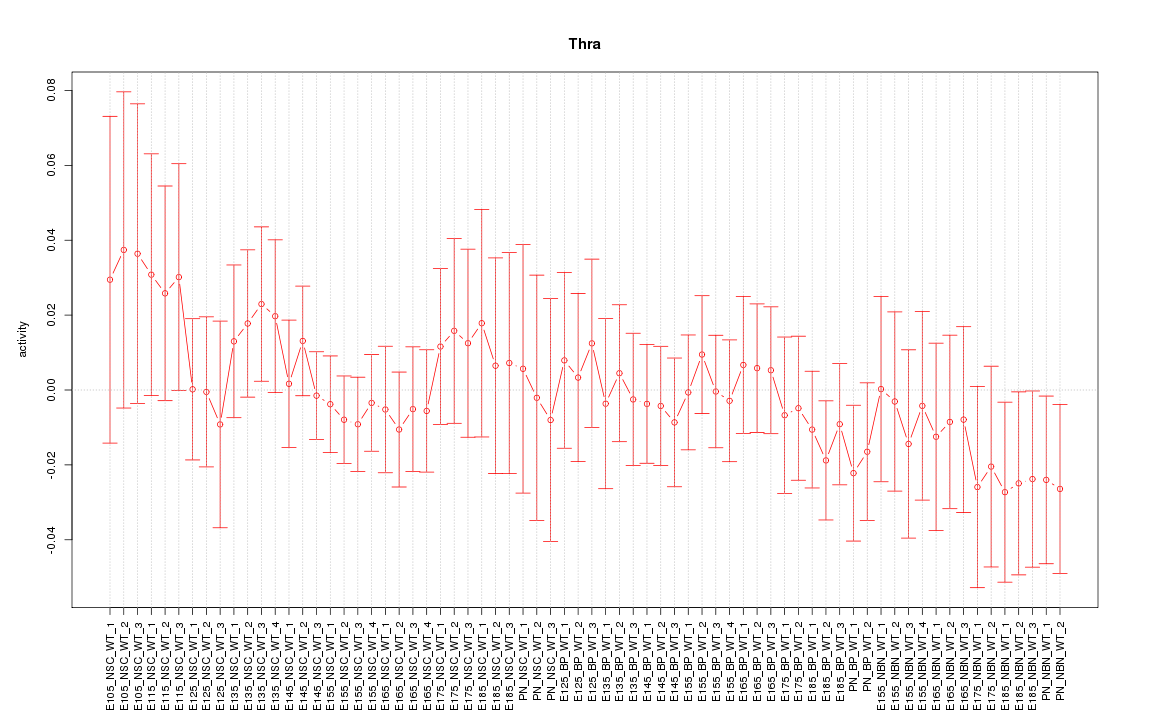
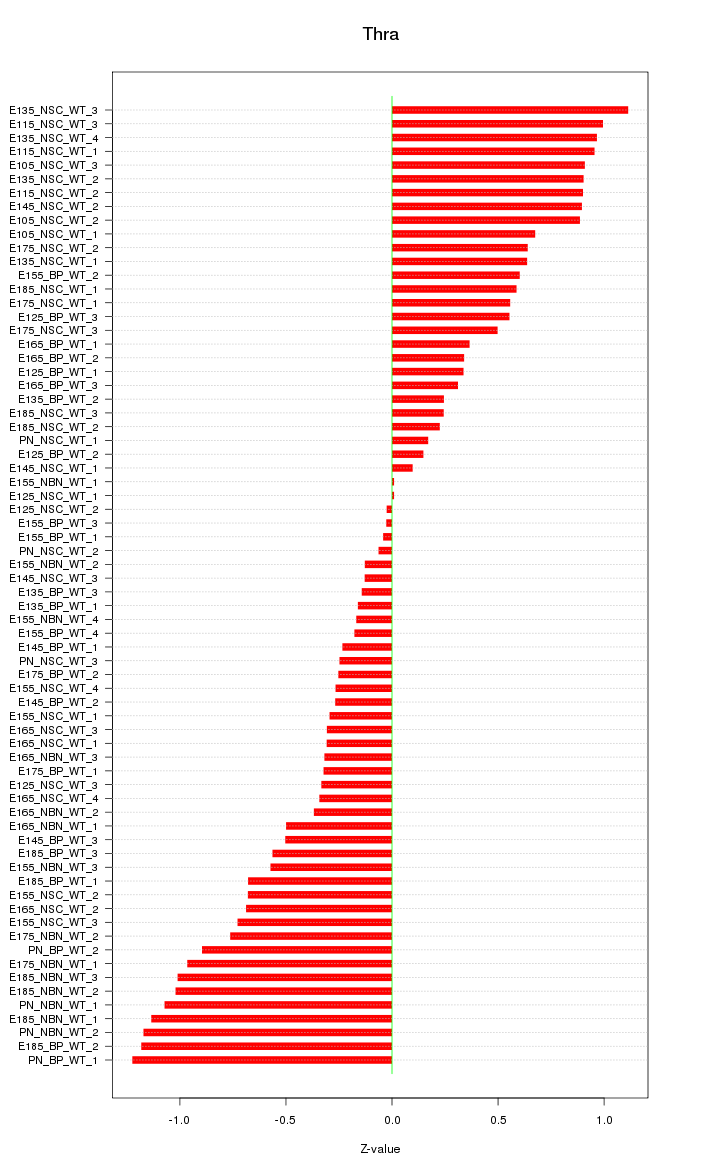
Motif ID: Thra
Z-value: 0.623
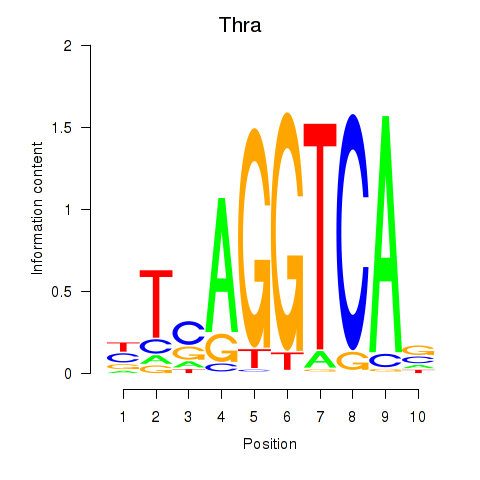
Transcription factors associated with Thra:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Thra | ENSMUSG00000058756.7 | Thra |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Thra | mm10_v2_chr11_+_98741805_98741816 | -0.82 | 4.7e-18 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.2 | GO:0097402 | neuroblast migration(GO:0097402) |
| 1.7 | 6.7 | GO:0098763 | mitotic cell cycle phase(GO:0098763) |
| 1.4 | 7.1 | GO:0014719 | skeletal muscle satellite cell activation(GO:0014719) |
| 0.9 | 5.2 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.8 | 2.5 | GO:1902915 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.8 | 3.1 | GO:0051295 | establishment of meiotic spindle localization(GO:0051295) |
| 0.7 | 2.2 | GO:0090202 | regulation of primitive erythrocyte differentiation(GO:0010725) transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.7 | 3.6 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.7 | 2.6 | GO:0071105 | response to interleukin-9(GO:0071104) response to interleukin-11(GO:0071105) |
| 0.7 | 4.6 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.6 | 1.9 | GO:0070093 | negative regulation of glucagon secretion(GO:0070093) |
| 0.6 | 1.8 | GO:0036118 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.5 | 1.6 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.5 | 3.5 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.4 | 4.3 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.4 | 5.9 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.4 | 1.7 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.4 | 2.9 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.4 | 1.2 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.4 | 1.2 | GO:0072554 | arterial endothelial cell fate commitment(GO:0060844) blood vessel endothelial cell fate commitment(GO:0060846) endothelial cell fate specification(GO:0060847) Notch signaling pathway involved in arterial endothelial cell fate commitment(GO:0060853) blood vessel lumenization(GO:0072554) blood vessel endothelial cell fate specification(GO:0097101) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) positive regulation of heart induction(GO:1901321) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.4 | 2.3 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.4 | 3.4 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.4 | 1.5 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.3 | 3.1 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.3 | 1.7 | GO:0051890 | regulation of cardioblast differentiation(GO:0051890) |
| 0.3 | 3.1 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.3 | 4.6 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.3 | 3.2 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.3 | 1.5 | GO:0034115 | negative regulation of heterotypic cell-cell adhesion(GO:0034115) cell-cell adhesion involved in gastrulation(GO:0070586) regulation of cell-cell adhesion involved in gastrulation(GO:0070587) negative regulation of protein kinase C signaling(GO:0090038) |
| 0.3 | 0.9 | GO:0002678 | positive regulation of chronic inflammatory response(GO:0002678) |
| 0.3 | 2.3 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.2 | 8.2 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.2 | 1.4 | GO:1903753 | cortical microtubule organization(GO:0043622) establishment of centrosome localization(GO:0051660) hard palate development(GO:0060022) negative regulation of p38MAPK cascade(GO:1903753) |
| 0.2 | 1.5 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.2 | 1.7 | GO:0061084 | negative regulation of protein refolding(GO:0061084) |
| 0.2 | 1.0 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.2 | 1.7 | GO:0019240 | citrulline biosynthetic process(GO:0019240) |
| 0.2 | 8.6 | GO:0008631 | intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0008631) |
| 0.2 | 2.8 | GO:0006337 | nucleosome disassembly(GO:0006337) protein-DNA complex disassembly(GO:0032986) |
| 0.2 | 2.0 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.2 | 0.5 | GO:0060854 | patterning of lymph vessels(GO:0060854) |
| 0.2 | 0.7 | GO:0035989 | tendon development(GO:0035989) |
| 0.2 | 1.2 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 0.2 | 1.0 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.2 | 5.0 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.2 | 1.1 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 | 0.4 | GO:1903795 | regulation of inorganic anion transmembrane transport(GO:1903795) |
| 0.1 | 2.0 | GO:0046628 | positive regulation of insulin receptor signaling pathway(GO:0046628) |
| 0.1 | 0.6 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.1 | 0.4 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.1 | 2.1 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.1 | 0.7 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.1 | 0.6 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 | 2.2 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 0.7 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.1 | 2.5 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.1 | 2.1 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 0.4 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.1 | 3.6 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.1 | 1.3 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 1.7 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 0.6 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.1 | 0.4 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.1 | 1.0 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 | 0.3 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) enamel mineralization(GO:0070166) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 0.4 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.1 | 0.3 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.1 | 1.6 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.6 | GO:0039703 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.0 | 0.9 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 5.2 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 1.3 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.0 | 0.3 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 3.7 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.2 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.6 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.2 | GO:0097494 | regulation of vesicle size(GO:0097494) |
| 0.0 | 2.8 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.7 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.5 | GO:0021702 | cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.4 | GO:0002385 | mucosal immune response(GO:0002385) |
| 0.0 | 0.1 | GO:0002741 | positive regulation of cytokine secretion involved in immune response(GO:0002741) |
| 0.0 | 1.1 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.6 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 0.2 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 0.3 | GO:0010388 | cullin deneddylation(GO:0010388) |
| 0.0 | 1.0 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 1.9 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 1.4 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 0.1 | GO:0045019 | negative regulation of nitric oxide biosynthetic process(GO:0045019) negative regulation of nitric oxide metabolic process(GO:1904406) |
| 0.0 | 0.2 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.0 | 2.0 | GO:0007498 | mesoderm development(GO:0007498) |
| 0.0 | 3.4 | GO:0060348 | bone development(GO:0060348) |
| 0.0 | 0.5 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.6 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.0 | 0.5 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.8 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.4 | GO:0031050 | dsRNA fragmentation(GO:0031050) production of miRNAs involved in gene silencing by miRNA(GO:0035196) production of small RNA involved in gene silencing by RNA(GO:0070918) |
| 0.0 | 0.4 | GO:0006760 | folic acid-containing compound metabolic process(GO:0006760) |
| 0.0 | 0.6 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.1 | GO:0042474 | middle ear morphogenesis(GO:0042474) |
| 0.0 | 0.3 | GO:1902108 | regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902108) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.6 | 3.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.5 | 5.0 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.4 | 8.8 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.3 | 5.0 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.3 | 7.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.3 | 1.4 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.3 | 1.0 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.2 | 1.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.2 | 5.2 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.2 | 1.6 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.2 | 4.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.2 | 0.6 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.2 | 0.6 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 1.9 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 2.8 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.1 | 1.6 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.1 | 2.0 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 1.3 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.1 | 3.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 0.9 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.1 | 3.7 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 2.1 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 0.7 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 1.8 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 0.7 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 1.2 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 1.7 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 4.6 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 5.9 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.4 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.4 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 3.7 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 2.1 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 2.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.4 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.6 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 1.7 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 2.2 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 2.0 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 1.9 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.8 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.6 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.9 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 1.4 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 3.5 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 1.0 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 2.1 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 0.5 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 5.8 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 1.7 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.4 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 3.9 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.6 | GO:0000118 | histone deacetylase complex(GO:0000118) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 5.0 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.6 | 1.8 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.6 | 2.3 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.5 | 3.8 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.5 | 3.6 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.4 | 4.3 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.4 | 3.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.4 | 5.2 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.3 | 2.6 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.3 | 1.6 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.3 | 1.7 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.3 | 4.6 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.3 | 5.2 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.3 | 1.4 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.3 | 0.5 | GO:0001224 | RNA polymerase II transcription cofactor binding(GO:0001224) |
| 0.2 | 1.9 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.2 | 1.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.2 | 3.6 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.2 | 3.7 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.2 | 0.8 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.2 | 8.0 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.2 | 0.9 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.2 | 2.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.2 | 3.1 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 2.2 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.2 | 1.2 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 1.0 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.6 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.1 | 4.3 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.1 | 1.0 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.6 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 3.5 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 0.4 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.1 | 1.9 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 0.5 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 0.2 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.1 | 0.4 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.1 | 0.8 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 1.3 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.1 | 0.7 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 3.6 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.1 | 0.2 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 0.4 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 2.1 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) |
| 0.0 | 2.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.7 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.2 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 1.4 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.2 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.6 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 1.2 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.9 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.9 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.7 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.0 | 0.6 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 2.9 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 1.6 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.4 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.9 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 1.1 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 1.0 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.0 | 1.4 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 10.1 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.5 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.7 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 1.0 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 6.3 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.3 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 1.2 | GO:0043851 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) rRNA (uridine-2'-O-)-methyltransferase activity(GO:0008650) rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) selenocysteine methyltransferase activity(GO:0016205) rRNA (adenine) methyltransferase activity(GO:0016433) rRNA (cytosine) methyltransferase activity(GO:0016434) rRNA (guanine) methyltransferase activity(GO:0016435) 1-phenanthrol methyltransferase activity(GO:0018707) protein-arginine N5-methyltransferase activity(GO:0019702) dimethylarsinite methyltransferase activity(GO:0034541) 4,5-dihydroxybenzo(a)pyrene methyltransferase activity(GO:0034807) 1-hydroxypyrene methyltransferase activity(GO:0034931) 1-hydroxy-6-methoxypyrene methyltransferase activity(GO:0034933) demethylmenaquinone methyltransferase activity(GO:0043770) cobalt-precorrin-6B C5-methyltransferase activity(GO:0043776) cobalt-precorrin-7 C15-methyltransferase activity(GO:0043777) cobalt-precorrin-5B C1-methyltransferase activity(GO:0043780) cobalt-precorrin-3 C17-methyltransferase activity(GO:0043782) dimethylamine methyltransferase activity(GO:0043791) hydroxyneurosporene-O-methyltransferase activity(GO:0043803) tRNA (adenine-57, 58-N(1)-) methyltransferase activity(GO:0043827) methylamine-specific methylcobalamin:coenzyme M methyltransferase activity(GO:0043833) trimethylamine methyltransferase activity(GO:0043834) methanol-specific methylcobalamin:coenzyme M methyltransferase activity(GO:0043851) monomethylamine methyltransferase activity(GO:0043852) P-methyltransferase activity(GO:0051994) Se-methyltransferase activity(GO:0051995) 2-phytyl-1,4-naphthoquinone methyltransferase activity(GO:0052624) tRNA (uracil-2'-O-)-methyltransferase activity(GO:0052665) tRNA (cytosine-2'-O-)-methyltransferase activity(GO:0052666) phosphomethylethanolamine N-methyltransferase activity(GO:0052667) tRNA (cytosine-3-)-methyltransferase activity(GO:0052735) rRNA (cytosine-2'-O-)-methyltransferase activity(GO:0070677) rRNA (cytosine-N4-)-methyltransferase activity(GO:0071424) trihydroxyferuloyl spermidine O-methyltransferase activity(GO:0080012) |
| 0.0 | 1.1 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 2.6 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.6 | GO:0008528 | G-protein coupled peptide receptor activity(GO:0008528) |
| 0.0 | 0.6 | GO:0020037 | heme binding(GO:0020037) |
| 0.0 | 1.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 2.3 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.2 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |