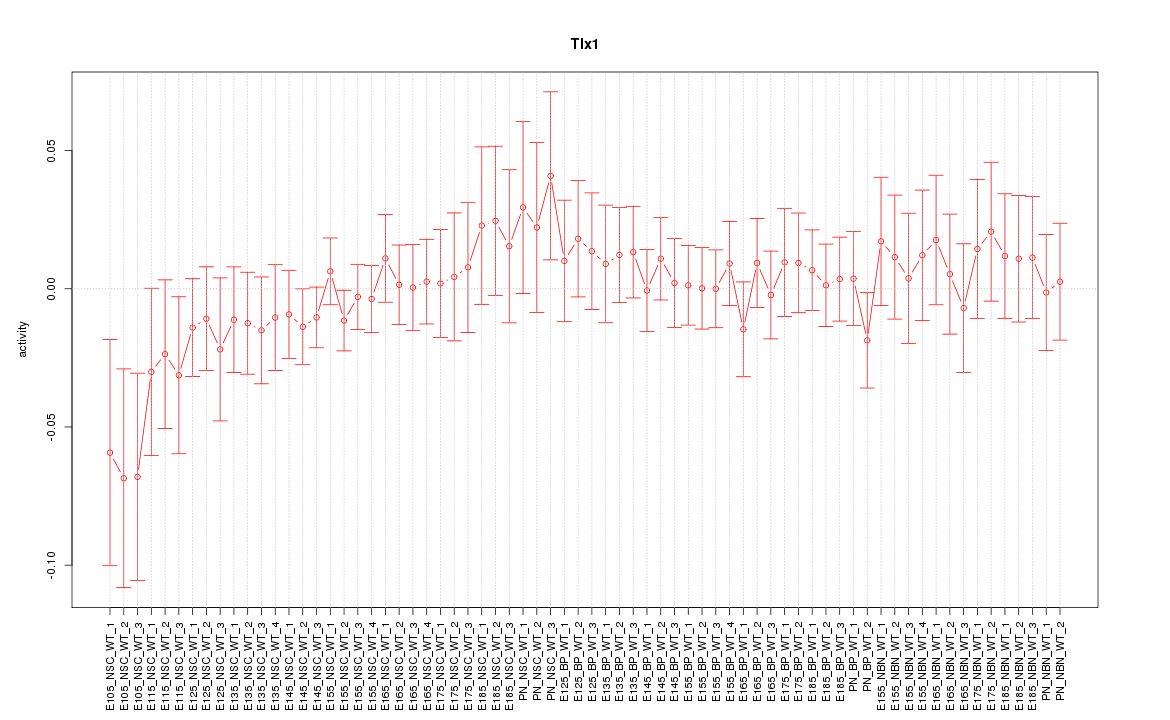
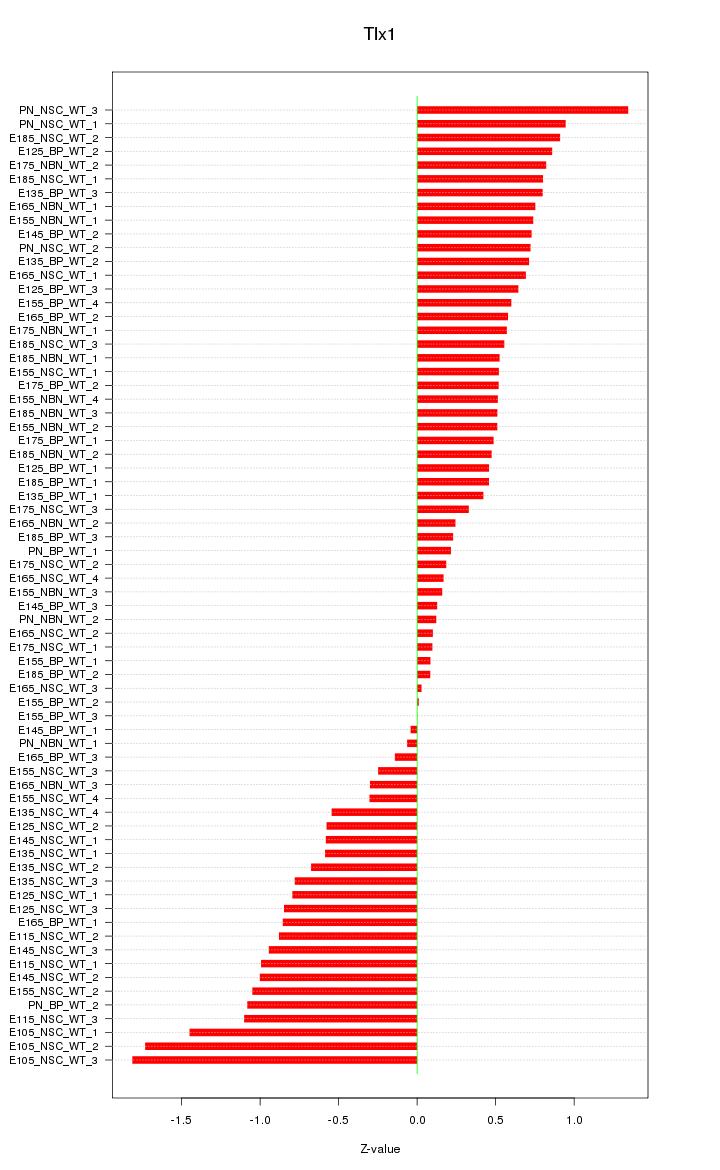
Motif ID: Tlx1
Z-value: 0.702
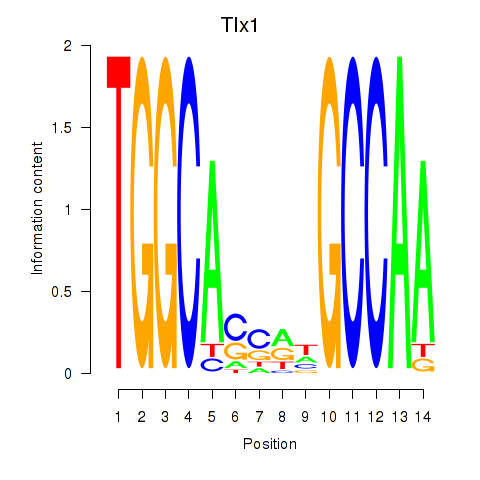
Transcription factors associated with Tlx1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Tlx1 | ENSMUSG00000025215.9 | Tlx1 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 9.4 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 1.5 | 4.6 | GO:0038095 | positive regulation of mast cell cytokine production(GO:0032765) Fc-epsilon receptor signaling pathway(GO:0038095) |
| 1.3 | 7.9 | GO:1902847 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of tau-protein kinase activity(GO:1902949) |
| 1.3 | 6.5 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 1.2 | 3.7 | GO:1904049 | regulation of spontaneous neurotransmitter secretion(GO:1904048) negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 1.1 | 4.5 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 1.0 | 6.1 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.9 | 7.2 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.9 | 4.4 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.8 | 9.6 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.7 | 2.2 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.7 | 3.5 | GO:0060050 | positive regulation of protein glycosylation(GO:0060050) |
| 0.7 | 6.0 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.7 | 2.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.6 | 5.1 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.6 | 1.9 | GO:0019401 | alditol biosynthetic process(GO:0019401) |
| 0.6 | 2.9 | GO:1902947 | regulation of tau-protein kinase activity(GO:1902947) |
| 0.6 | 3.9 | GO:0015840 | urea transport(GO:0015840) urea transmembrane transport(GO:0071918) |
| 0.5 | 2.7 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.5 | 1.5 | GO:0050883 | musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.4 | 2.2 | GO:0051012 | microtubule sliding(GO:0051012) negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 0.4 | 2.6 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.4 | 1.3 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.4 | 0.9 | GO:0045963 | negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) |
| 0.4 | 1.3 | GO:0051933 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) L-glutamate(1-) import into cell(GO:1903802) L-glutamate import into cell(GO:1990123) |
| 0.4 | 1.6 | GO:0046950 | cellular ketone body metabolic process(GO:0046950) |
| 0.4 | 4.1 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.4 | 2.2 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.4 | 1.8 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.3 | 1.0 | GO:0034035 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) |
| 0.3 | 3.2 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.3 | 2.2 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.3 | 0.9 | GO:0030070 | insulin processing(GO:0030070) |
| 0.3 | 0.9 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 0.3 | 2.7 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.3 | 3.4 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.3 | 1.4 | GO:0010359 | regulation of anion channel activity(GO:0010359) |
| 0.3 | 1.3 | GO:0032911 | nerve growth factor production(GO:0032902) negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.3 | 2.6 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.2 | 0.7 | GO:0032430 | positive regulation of phospholipase A2 activity(GO:0032430) |
| 0.2 | 3.4 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.2 | 1.3 | GO:2000664 | positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.2 | 1.7 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.2 | 3.7 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.2 | 0.6 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.2 | 1.0 | GO:0071476 | cellular hypotonic response(GO:0071476) |
| 0.2 | 0.7 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.2 | 2.9 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.2 | 1.2 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.2 | 1.5 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 0.2 | 4.9 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) |
| 0.2 | 1.1 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.2 | 0.5 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.2 | 1.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.2 | 1.8 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.1 | 2.4 | GO:0010640 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) |
| 0.1 | 4.4 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.1 | 3.0 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 0.6 | GO:1903598 | negative regulation of pinocytosis(GO:0048550) negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) positive regulation of gap junction assembly(GO:1903598) |
| 0.1 | 0.8 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 2.6 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 1.8 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 1.2 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 0.4 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.1 | 1.4 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.1 | 0.3 | GO:0070488 | neutrophil aggregation(GO:0070488) |
| 0.1 | 1.1 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.1 | 3.4 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 3.2 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.1 | 2.4 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.1 | 2.9 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 4.0 | GO:0048678 | response to axon injury(GO:0048678) |
| 0.1 | 0.6 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.1 | 1.5 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.1 | 1.2 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.1 | 0.3 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.1 | 2.1 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 0.6 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 | 8.8 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.1 | 0.6 | GO:0071225 | cellular response to muramyl dipeptide(GO:0071225) |
| 0.0 | 3.6 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.1 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.0 | 1.8 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.1 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.9 | GO:0002437 | inflammatory response to antigenic stimulus(GO:0002437) |
| 0.0 | 3.1 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.6 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 2.5 | GO:0001701 | in utero embryonic development(GO:0001701) |
| 0.0 | 0.5 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.2 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.5 | GO:0030307 | positive regulation of cell growth(GO:0030307) |
| 0.0 | 0.1 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 1.4 | GO:0006024 | glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.8 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 3.3 | GO:0030509 | BMP signaling pathway(GO:0030509) |
| 0.0 | 2.7 | GO:0098792 | xenophagy(GO:0098792) |
| 0.0 | 2.9 | GO:0007612 | learning(GO:0007612) |
| 0.0 | 0.0 | GO:0010455 | positive regulation of cell fate commitment(GO:0010455) |
| 0.0 | 0.1 | GO:0070944 | neutrophil mediated killing of bacterium(GO:0070944) |
| 0.0 | 0.4 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.4 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 4.0 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.1 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 1.4 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 2.2 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.4 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 0.1 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.2 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 7.9 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) neurofibrillary tangle(GO:0097418) |
| 0.7 | 3.7 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.4 | 2.0 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.3 | 0.9 | GO:0043512 | inhibin A complex(GO:0043512) |
| 0.3 | 0.8 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.3 | 18.6 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.2 | 0.7 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.2 | 2.7 | GO:0031045 | dense core granule(GO:0031045) |
| 0.2 | 6.6 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 0.4 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.1 | 0.5 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.1 | 2.9 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 1.3 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.1 | 2.1 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 6.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 4.1 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 1.7 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 1.8 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 1.3 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.1 | 0.6 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.1 | 1.4 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.1 | 4.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 0.8 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.2 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 1.2 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 4.8 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.1 | 5.4 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 2.8 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.0 | 2.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 5.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.6 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 1.8 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 2.4 | GO:0032589 | neuron projection membrane(GO:0032589) |
| 0.0 | 2.2 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.2 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 1.2 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 1.5 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.7 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 5.3 | GO:0030017 | sarcomere(GO:0030017) |
| 0.0 | 0.8 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 2.6 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 3.4 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 1.0 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.2 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 1.2 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 1.9 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 1.7 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.4 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 1.3 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 3.2 | GO:0005929 | cilium(GO:0005929) |
| 0.0 | 3.9 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 3.2 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 2.2 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.8 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 4.3 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 0.1 | GO:0097361 | CIA complex(GO:0097361) |
| 0.0 | 1.2 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.6 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 2.3 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.9 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 9.6 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 1.6 | 6.5 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 1.1 | 4.4 | GO:0004096 | catalase activity(GO:0004096) |
| 1.1 | 3.3 | GO:0008521 | acetyl-CoA transporter activity(GO:0008521) |
| 0.9 | 3.5 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.8 | 4.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.8 | 6.2 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.7 | 9.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.6 | 3.9 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.6 | 3.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.6 | 3.7 | GO:0016453 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.5 | 2.2 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.5 | 1.9 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 0.4 | 1.3 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.4 | 3.0 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.4 | 1.3 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.4 | 1.6 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.4 | 2.4 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.3 | 7.9 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.3 | 4.6 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.3 | 0.9 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.3 | 1.1 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.3 | 6.1 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.3 | 5.5 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.3 | 1.4 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.3 | 3.0 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.3 | 5.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.2 | 2.9 | GO:0015279 | store-operated calcium channel activity(GO:0015279) inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.2 | 1.6 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.2 | 1.6 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.2 | 1.7 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.2 | 0.6 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 0.2 | 0.8 | GO:0050347 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.2 | 5.6 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.2 | 5.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.2 | 2.0 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.2 | 1.3 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.2 | 1.0 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.2 | 0.9 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.2 | 0.9 | GO:0070191 | methionine-R-sulfoxide reductase activity(GO:0070191) |
| 0.1 | 2.6 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 1.0 | GO:0004779 | sulfate adenylyltransferase activity(GO:0004779) |
| 0.1 | 0.6 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 0.6 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.1 | 0.8 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 1.5 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 6.9 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 1.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 2.6 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 0.4 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.9 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 3.4 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 5.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 1.8 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 2.0 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.1 | 3.6 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 1.7 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 0.3 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.1 | 2.1 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 1.3 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) |
| 0.0 | 0.1 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 2.3 | GO:0044824 | integrase activity(GO:0008907) T/G mismatch-specific endonuclease activity(GO:0043765) retroviral integrase activity(GO:0044823) retroviral 3' processing activity(GO:0044824) |
| 0.0 | 1.6 | GO:0008748 | N-ethylmaleimide reductase activity(GO:0008748) reduced coenzyme F420 dehydrogenase activity(GO:0043738) sulfur oxygenase reductase activity(GO:0043826) malolactic enzyme activity(GO:0043883) epoxyqueuosine reductase activity(GO:0052693) |
| 0.0 | 0.8 | GO:0035380 | C-3 sterol dehydrogenase (C-4 sterol decarboxylase) activity(GO:0000252) mevaldate reductase activity(GO:0004495) gluconate dehydrogenase activity(GO:0008875) epoxide dehydrogenase activity(GO:0018451) 5-exo-hydroxycamphor dehydrogenase activity(GO:0018452) 2-hydroxytetrahydrofuran dehydrogenase activity(GO:0018453) acetoin dehydrogenase activity(GO:0019152) phenylcoumaran benzylic ether reductase activity(GO:0032442) D-xylose:NADP reductase activity(GO:0032866) L-arabinose:NADP reductase activity(GO:0032867) D-arabinitol dehydrogenase, D-ribulose forming (NADP+) activity(GO:0033709) (R)-(-)-1,2,3,4-tetrahydronaphthol dehydrogenase activity(GO:0034831) 3-hydroxymenthone dehydrogenase activity(GO:0034840) very long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0035380) dihydrotestosterone 17-beta-dehydrogenase activity(GO:0035410) (R)-2-hydroxyisocaproate dehydrogenase activity(GO:0043713) L-arabinose 1-dehydrogenase (NADP+) activity(GO:0044103) L-xylulose reductase (NAD+) activity(GO:0044105) 3-ketoglucose-reductase activity(GO:0048258) D-arabinitol dehydrogenase, D-xylulose forming (NADP+) activity(GO:0052677) |
| 0.0 | 1.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 1.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 1.6 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.5 | GO:0050253 | prenylcysteine methylesterase activity(GO:0010296) 1-oxa-2-oxocycloheptane lactonase activity(GO:0018731) sulfolactone hydrolase activity(GO:0018732) butyrolactone hydrolase activity(GO:0018734) endosulfan lactone lactonase activity(GO:0034892) L-ascorbate 6-phosphate lactonase activity(GO:0035460) Ser-tRNA(Thr) hydrolase activity(GO:0043905) Ala-tRNA(Pro) hydrolase activity(GO:0043906) Cys-tRNA(Pro) hydrolase activity(GO:0043907) Ser(Gly)-tRNA(Ala) hydrolase activity(GO:0043908) all-trans-retinyl-palmitate hydrolase, all-trans-retinol forming activity(GO:0047376) retinyl-palmitate esterase activity(GO:0050253) mannosyl-oligosaccharide 1,6-alpha-mannosidase activity(GO:0052767) mannosyl-oligosaccharide 1,3-alpha-mannosidase activity(GO:0052768) methyl indole-3-acetate esterase activity(GO:0080030) methyl salicylate esterase activity(GO:0080031) methyl jasmonate esterase activity(GO:0080032) |
| 0.0 | 0.6 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 1.4 | GO:0035496 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) O antigen polymerase activity(GO:0008755) lipopolysaccharide-1,6-galactosyltransferase activity(GO:0008921) cellulose synthase activity(GO:0016759) 9-phenanthrol UDP-glucuronosyltransferase activity(GO:0018715) 1-phenanthrol glycosyltransferase activity(GO:0018716) 9-phenanthrol glycosyltransferase activity(GO:0018717) 1,2-dihydroxy-phenanthrene glycosyltransferase activity(GO:0018718) phenanthrol glycosyltransferase activity(GO:0019112) alpha-1,2-galactosyltransferase activity(GO:0031278) dolichyl pyrophosphate Man7GlcNAc2 alpha-1,3-glucosyltransferase activity(GO:0033556) endogalactosaminidase activity(GO:0033931) lipopolysaccharide-1,5-galactosyltransferase activity(GO:0035496) dolichyl pyrophosphate Glc1Man9GlcNAc2 alpha-1,3-glucosyltransferase activity(GO:0042283) inositol phosphoceramide synthase activity(GO:0045140) alpha-(1->3)-fucosyltransferase activity(GO:0046920) indole-3-butyrate beta-glucosyltransferase activity(GO:0052638) salicylic acid glucosyltransferase (ester-forming) activity(GO:0052639) salicylic acid glucosyltransferase (glucoside-forming) activity(GO:0052640) benzoic acid glucosyltransferase activity(GO:0052641) chondroitin hydrolase activity(GO:0052757) dolichyl-pyrophosphate Man7GlcNAc2 alpha-1,6-mannosyltransferase activity(GO:0052824) cytokinin 9-beta-glucosyltransferase activity(GO:0080062) |
| 0.0 | 3.1 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.0 | 1.2 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.2 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 2.7 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 2.1 | GO:0004713 | protein tyrosine kinase activity(GO:0004713) |
| 0.0 | 0.5 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.7 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.8 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 1.6 | GO:0016247 | channel regulator activity(GO:0016247) |
| 0.0 | 0.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |