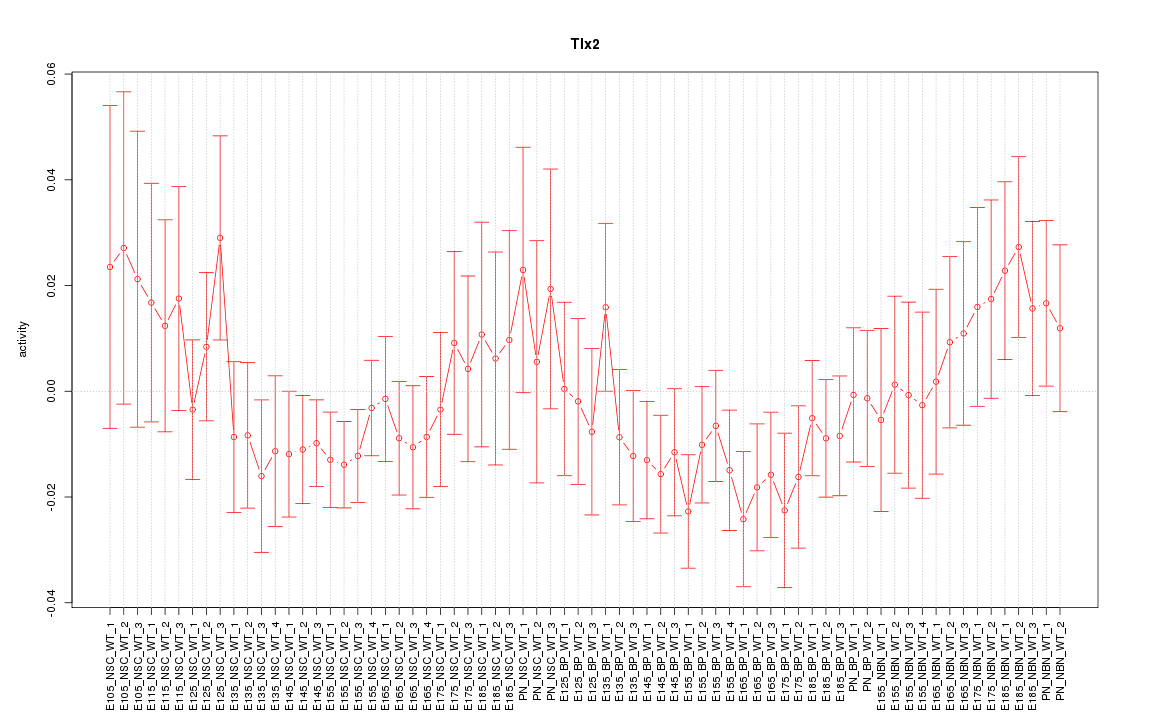
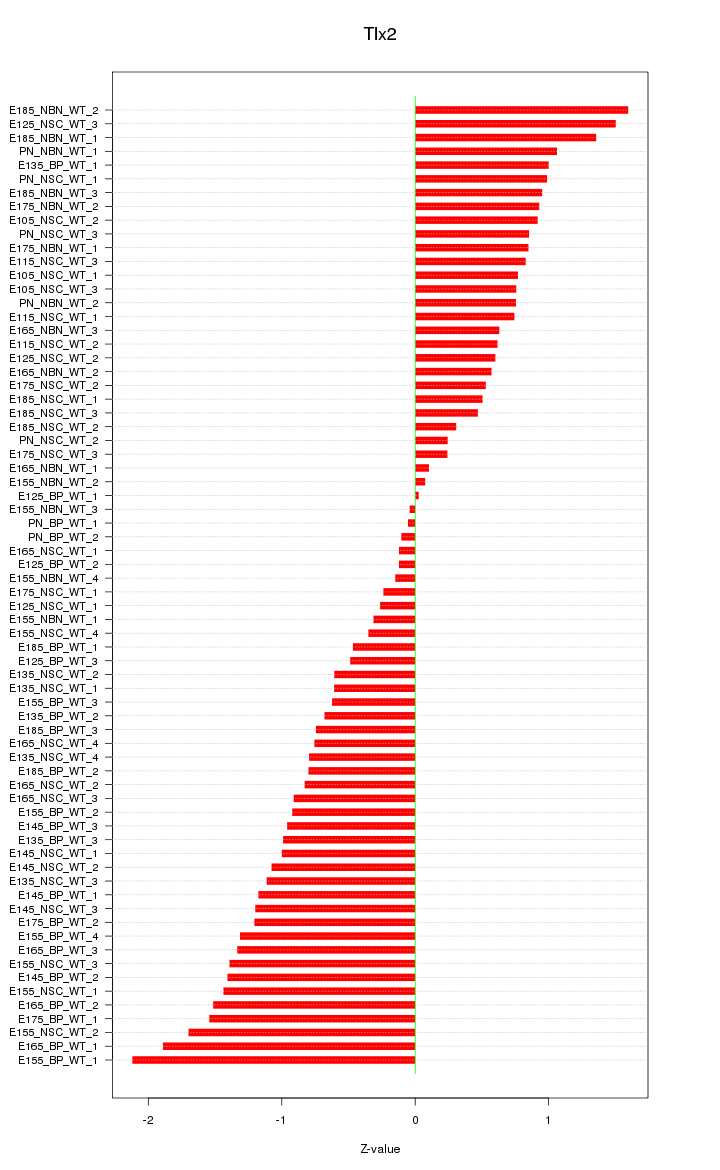
Motif ID: Tlx2
Z-value: 0.934
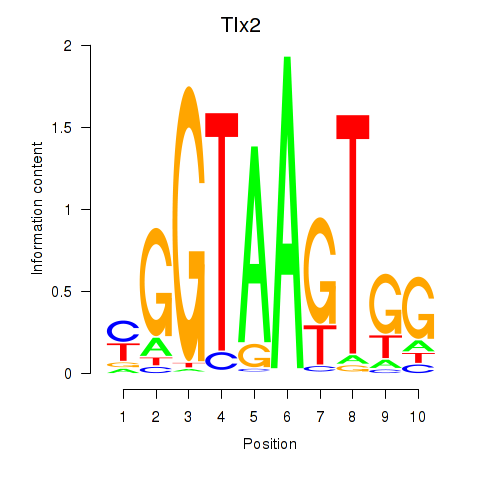
Transcription factors associated with Tlx2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Tlx2 | ENSMUSG00000068327.3 | Tlx2 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 10.3 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 2.3 | 7.0 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 1.6 | 4.8 | GO:0003278 | apoptotic process involved in heart morphogenesis(GO:0003278) |
| 1.3 | 3.8 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 1.2 | 3.7 | GO:1903048 | regulation of acetylcholine-gated cation channel activity(GO:1903048) |
| 1.2 | 4.7 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 1.0 | 4.1 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.9 | 2.8 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 0.9 | 2.7 | GO:0090187 | zymogen granule exocytosis(GO:0070625) positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.8 | 2.5 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.8 | 3.2 | GO:2000974 | negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.8 | 2.4 | GO:0097461 | ferric iron import(GO:0033216) ferric iron import into cell(GO:0097461) |
| 0.8 | 2.3 | GO:1900158 | negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.7 | 2.9 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.7 | 4.3 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.7 | 2.0 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.6 | 1.8 | GO:0006296 | base-excision repair, AP site formation(GO:0006285) nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.6 | 1.7 | GO:2000562 | negative regulation of interferon-gamma secretion(GO:1902714) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.5 | 1.6 | GO:0010752 | signal complex assembly(GO:0007172) regulation of cGMP-mediated signaling(GO:0010752) |
| 0.5 | 1.6 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.5 | 3.1 | GO:0036233 | glycine import(GO:0036233) |
| 0.5 | 0.5 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.5 | 1.5 | GO:0001803 | antibody-dependent cellular cytotoxicity(GO:0001788) type III hypersensitivity(GO:0001802) regulation of type III hypersensitivity(GO:0001803) positive regulation of type III hypersensitivity(GO:0001805) |
| 0.5 | 1.9 | GO:0002339 | B cell selection(GO:0002339) |
| 0.5 | 1.4 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.5 | 3.7 | GO:0002681 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.4 | 4.1 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.4 | 1.2 | GO:0030300 | regulation of intestinal cholesterol absorption(GO:0030300) |
| 0.4 | 3.2 | GO:0046645 | positive regulation of gamma-delta T cell differentiation(GO:0045588) positive regulation of gamma-delta T cell activation(GO:0046645) positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.4 | 1.6 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.4 | 3.0 | GO:0032725 | positive regulation of granulocyte macrophage colony-stimulating factor production(GO:0032725) |
| 0.4 | 1.1 | GO:0007521 | muscle cell fate determination(GO:0007521) mammary placode formation(GO:0060596) |
| 0.4 | 1.1 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.4 | 1.1 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) terminal web assembly(GO:1902896) |
| 0.4 | 1.1 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.4 | 1.1 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.3 | 0.7 | GO:0071649 | regulation of chemokine (C-C motif) ligand 5 production(GO:0071649) |
| 0.3 | 3.0 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.3 | 2.3 | GO:0007343 | egg activation(GO:0007343) |
| 0.3 | 1.0 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.3 | 2.9 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.3 | 0.9 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.3 | 3.1 | GO:0009186 | deoxyribonucleoside diphosphate metabolic process(GO:0009186) |
| 0.3 | 1.8 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.3 | 1.4 | GO:0007320 | insemination(GO:0007320) |
| 0.3 | 1.1 | GO:0006842 | tricarboxylic acid transport(GO:0006842) succinate transport(GO:0015744) citrate transport(GO:0015746) |
| 0.3 | 1.1 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.3 | 3.5 | GO:0031017 | exocrine pancreas development(GO:0031017) |
| 0.2 | 1.7 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.2 | 2.6 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.2 | 0.7 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.2 | 2.3 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.2 | 0.4 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.2 | 5.5 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.2 | 0.6 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.2 | 0.8 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 0.2 | 1.4 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.2 | 5.4 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.2 | 2.2 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) sphingoid biosynthetic process(GO:0046520) |
| 0.2 | 0.6 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.2 | 0.6 | GO:0010511 | serotonin receptor signaling pathway(GO:0007210) regulation of phosphatidylinositol biosynthetic process(GO:0010511) |
| 0.2 | 0.8 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.2 | 0.9 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.2 | 1.1 | GO:0097491 | trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.2 | 0.7 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 0.2 | 0.7 | GO:0036518 | chemorepulsion of dopaminergic neuron axon(GO:0036518) |
| 0.2 | 1.4 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.2 | 0.7 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.2 | 0.5 | GO:0021577 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.2 | 2.2 | GO:0060586 | multicellular organismal iron ion homeostasis(GO:0060586) |
| 0.2 | 1.8 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.2 | 1.1 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.2 | 0.8 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.2 | 0.8 | GO:0045918 | negative regulation of cytolysis(GO:0045918) |
| 0.2 | 1.4 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.2 | 1.4 | GO:0045056 | transcytosis(GO:0045056) |
| 0.2 | 2.3 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.1 | 0.9 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.1 | 0.9 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.4 | GO:0048352 | neural plate mediolateral regionalization(GO:0021998) mesoderm structural organization(GO:0048338) paraxial mesoderm structural organization(GO:0048352) |
| 0.1 | 3.1 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.1 | 0.4 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 | 0.4 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.1 | 2.2 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 1.5 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 2.6 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.1 | 1.1 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.1 | 0.7 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 | 0.2 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.1 | 0.2 | GO:1904948 | midbrain dopaminergic neuron differentiation(GO:1904948) |
| 0.1 | 0.8 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.1 | 2.0 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 3.1 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 0.3 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.1 | 0.4 | GO:0097298 | regulation of nucleus size(GO:0097298) |
| 0.1 | 1.5 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 2.0 | GO:0001516 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.1 | 0.3 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.1 | 0.5 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.1 | 1.2 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.1 | 1.0 | GO:0046036 | GTP biosynthetic process(GO:0006183) CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.1 | 1.3 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.1 | 1.8 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.1 | 1.9 | GO:0033622 | integrin activation(GO:0033622) |
| 0.1 | 2.2 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.1 | 0.2 | GO:0051342 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.1 | 3.9 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.1 | 0.9 | GO:0042104 | positive regulation of activated T cell proliferation(GO:0042104) |
| 0.1 | 2.2 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.1 | 0.3 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.1 | 0.3 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.1 | 1.7 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 2.3 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 0.7 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.1 | 0.2 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.1 | 0.6 | GO:0007100 | mitotic centrosome separation(GO:0007100) |
| 0.1 | 1.1 | GO:0045026 | plasma membrane fusion(GO:0045026) |
| 0.1 | 2.2 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.1 | 1.1 | GO:0018126 | protein hydroxylation(GO:0018126) |
| 0.1 | 3.4 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.1 | 1.3 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 0.3 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.7 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.1 | 0.6 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 0.5 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.1 | 0.7 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 1.3 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.1 | 0.9 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 | 0.6 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.1 | 0.6 | GO:0008637 | apoptotic mitochondrial changes(GO:0008637) |
| 0.1 | 0.2 | GO:0032512 | regulation of protein phosphatase type 2B activity(GO:0032512) negative regulation of protein phosphatase type 2B activity(GO:0032513) |
| 0.1 | 0.3 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.1 | 0.4 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.1 | 0.5 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 0.6 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 0.3 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.1 | 0.2 | GO:1904177 | regulation of adipose tissue development(GO:1904177) |
| 0.1 | 1.3 | GO:1903861 | regulation of dendrite extension(GO:1903859) positive regulation of dendrite extension(GO:1903861) |
| 0.1 | 0.2 | GO:0051775 | response to redox state(GO:0051775) |
| 0.1 | 0.2 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.1 | 1.0 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.1 | 1.2 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.1 | 0.4 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 1.1 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.1 | 1.2 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.1 | 0.2 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) |
| 0.1 | 0.4 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.1 | 0.9 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.1 | 0.4 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 | 0.7 | GO:0061000 | negative regulation of dendritic spine development(GO:0061000) |
| 0.1 | 1.8 | GO:0007099 | centriole replication(GO:0007099) |
| 0.1 | 0.2 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.1 | 0.7 | GO:0010388 | cullin deneddylation(GO:0010388) |
| 0.0 | 0.2 | GO:2000383 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.0 | 0.2 | GO:0043090 | amino acid import(GO:0043090) |
| 0.0 | 0.1 | GO:0070488 | neutrophil aggregation(GO:0070488) |
| 0.0 | 1.7 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.8 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 | 0.9 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.0 | 1.5 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 1.5 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 1.8 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 1.9 | GO:0014911 | positive regulation of smooth muscle cell migration(GO:0014911) |
| 0.0 | 1.6 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.0 | 0.4 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 1.0 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.3 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 2.1 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.0 | 0.2 | GO:0098734 | protein depalmitoylation(GO:0002084) negative regulation of Golgi to plasma membrane protein transport(GO:0042997) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 1.1 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 1.9 | GO:0035058 | nonmotile primary cilium assembly(GO:0035058) |
| 0.0 | 0.1 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 0.0 | 0.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.9 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.3 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.0 | 0.1 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.0 | 0.2 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.5 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 0.1 | GO:0070844 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.0 | 0.1 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.4 | GO:0033262 | regulation of nuclear cell cycle DNA replication(GO:0033262) |
| 0.0 | 0.1 | GO:0051176 | negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) positive regulation of sulfur metabolic process(GO:0051176) |
| 0.0 | 0.8 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.0 | 2.7 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.2 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.1 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.0 | 0.2 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.7 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 1.2 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.0 | 0.6 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.0 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.0 | 0.8 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.5 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.4 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.0 | 0.1 | GO:0034397 | telomere localization(GO:0034397) |
| 0.0 | 0.7 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.2 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.3 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.4 | GO:0043403 | skeletal muscle tissue regeneration(GO:0043403) |
| 0.0 | 0.9 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.2 | GO:0002093 | auditory receptor cell morphogenesis(GO:0002093) |
| 0.0 | 0.9 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.6 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.4 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.2 | GO:1903301 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.9 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 1.9 | GO:0000082 | G1/S transition of mitotic cell cycle(GO:0000082) |
| 0.0 | 0.3 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.7 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.7 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.1 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.1 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.1 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.3 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.5 | GO:0043627 | response to estrogen(GO:0043627) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.0 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.5 | 2.3 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.4 | 1.8 | GO:0071942 | XPC complex(GO:0071942) |
| 0.4 | 4.3 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.4 | 1.2 | GO:0005940 | septin ring(GO:0005940) |
| 0.4 | 1.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.3 | 3.7 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.3 | 1.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.3 | 4.1 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.3 | 4.7 | GO:0043205 | fibril(GO:0043205) |
| 0.3 | 1.9 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 2.7 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.2 | 2.3 | GO:0098533 | ATPase dependent transmembrane transport complex(GO:0098533) |
| 0.2 | 1.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.2 | 1.1 | GO:0002177 | manchette(GO:0002177) |
| 0.2 | 0.6 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.2 | 1.5 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.2 | 1.7 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.2 | 2.3 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.2 | 4.2 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.2 | 0.9 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.2 | 1.9 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.2 | 0.5 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.2 | 1.4 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 0.4 | GO:0055087 | Ski complex(GO:0055087) |
| 0.1 | 2.8 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 2.1 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.1 | 0.7 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.1 | 1.0 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 2.8 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 8.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 2.4 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 1.1 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.1 | 0.7 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.1 | 0.9 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 0.1 | 0.5 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.1 | 1.6 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 1.6 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 1.9 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 0.5 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 1.1 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 0.8 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 0.8 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 1.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 2.6 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 0.3 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 0.5 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 2.9 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 0.6 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 4.2 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 0.5 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 1.0 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 0.7 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.3 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.1 | 0.8 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 0.3 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.7 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 0.2 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.0 | 1.3 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 2.5 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.6 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.6 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.0 | 1.5 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 1.6 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 2.7 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.3 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.4 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.5 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.6 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 1.5 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 2.2 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.7 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 1.7 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 4.6 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 2.3 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.7 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 3.6 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 3.0 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.2 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.1 | GO:0045240 | dihydrolipoyl dehydrogenase complex(GO:0045240) |
| 0.0 | 5.6 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 1.0 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.8 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.8 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 5.2 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 1.2 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.4 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 10.5 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 1.0 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.2 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 1.7 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.4 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.2 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.7 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.2 | GO:0044447 | axoneme part(GO:0044447) |
| 0.0 | 0.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.1 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.3 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.2 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) cytosolic proteasome complex(GO:0031597) |
| 0.0 | 2.9 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.7 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 0.2 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 1.1 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.6 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 2.8 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.8 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.2 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.5 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 11.9 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 0.9 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.1 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 1.4 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.1 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 4.7 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.9 | 5.4 | GO:0043426 | MRF binding(GO:0043426) |
| 0.8 | 2.4 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.7 | 4.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.6 | 3.2 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.6 | 1.9 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 0.6 | 3.0 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.6 | 2.3 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) prostaglandin-E synthase activity(GO:0050220) |
| 0.6 | 2.9 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.5 | 2.2 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.5 | 2.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.5 | 3.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.5 | 1.5 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.4 | 1.1 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.4 | 1.1 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.4 | 1.1 | GO:0031687 | A2A adenosine receptor binding(GO:0031687) |
| 0.4 | 1.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.4 | 2.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.4 | 1.4 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.3 | 3.5 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.3 | 2.7 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.3 | 3.7 | GO:0004889 | acetylcholine-activated cation-selective channel activity(GO:0004889) |
| 0.3 | 4.4 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.3 | 2.2 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.3 | 1.7 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.3 | 6.0 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.3 | 1.9 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.2 | 1.7 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.2 | 1.9 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.2 | 0.7 | GO:0030362 | protein phosphatase type 4 regulator activity(GO:0030362) |
| 0.2 | 1.6 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.2 | 1.4 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.2 | 2.2 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.2 | 7.0 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.2 | 0.4 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.2 | 1.9 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.2 | 0.6 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.2 | 0.6 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) glycoprotein transporter activity(GO:0034437) |
| 0.2 | 2.3 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.2 | 0.6 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin binding(GO:0051378) serotonin receptor activity(GO:0099589) |
| 0.2 | 0.8 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.2 | 0.9 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.2 | 0.5 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.2 | 1.4 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.2 | 0.7 | GO:0035473 | lipase binding(GO:0035473) |
| 0.2 | 0.9 | GO:0016615 | malate dehydrogenase activity(GO:0016615) |
| 0.2 | 0.7 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.2 | 1.4 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.2 | 1.8 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.2 | 1.7 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 4.1 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 0.7 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.1 | 1.9 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.1 | 2.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 1.1 | GO:0103116 | alpha-D-galactofuranose transporter activity(GO:0103116) steroid hormone binding(GO:1990239) |
| 0.1 | 1.8 | GO:0070636 | single-strand selective uracil DNA N-glycosylase activity(GO:0017065) nicotinamide riboside hydrolase activity(GO:0070635) nicotinic acid riboside hydrolase activity(GO:0070636) deoxyribonucleoside 5'-monophosphate N-glycosidase activity(GO:0070694) |
| 0.1 | 0.7 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.1 | 1.8 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) acidic amino acid transmembrane transporter activity(GO:0015172) |
| 0.1 | 0.8 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 0.8 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.1 | 0.4 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.1 | 0.4 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.1 | 0.4 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.1 | 0.7 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 1.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 1.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.1 | 0.3 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.1 | 1.5 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 0.9 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.1 | 2.5 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) |
| 0.1 | 1.1 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 1.4 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.1 | 4.9 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 0.9 | GO:0015279 | store-operated calcium channel activity(GO:0015279) inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.1 | 0.7 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 1.6 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 0.9 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.3 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.1 | 0.8 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.1 | 0.5 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.2 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 1.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 2.1 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 2.1 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 1.0 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 1.0 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.1 | 0.7 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 3.2 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.1 | 0.4 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.1 | 1.1 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.1 | 0.2 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 0.7 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 8.5 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 0.2 | GO:0036004 | GAF domain binding(GO:0036004) |
| 0.1 | 0.7 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 6.7 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 0.5 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 1.0 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 0.9 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 1.9 | GO:0050694 | galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 0.3 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.1 | 1.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.8 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 0.2 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 0.5 | GO:0052872 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) phenanthrene 9,10-monooxygenase activity(GO:0018636) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) tocotrienol omega-hydroxylase activity(GO:0052872) thalianol hydroxylase activity(GO:0080014) |
| 0.0 | 1.3 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.8 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 1.8 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 11.4 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 1.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.5 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 4.3 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.2 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 2.5 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 0.7 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 1.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.2 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.6 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.6 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.5 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.6 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.8 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 1.8 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 3.0 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 1.2 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.8 | GO:0019707 | protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 5.1 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.7 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 1.9 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.1 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.2 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 7.7 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.9 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.0 | 0.3 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.2 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.5 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.2 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 2.6 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 0.2 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.4 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.7 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.5 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.8 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.0 | 0.9 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 0.4 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |