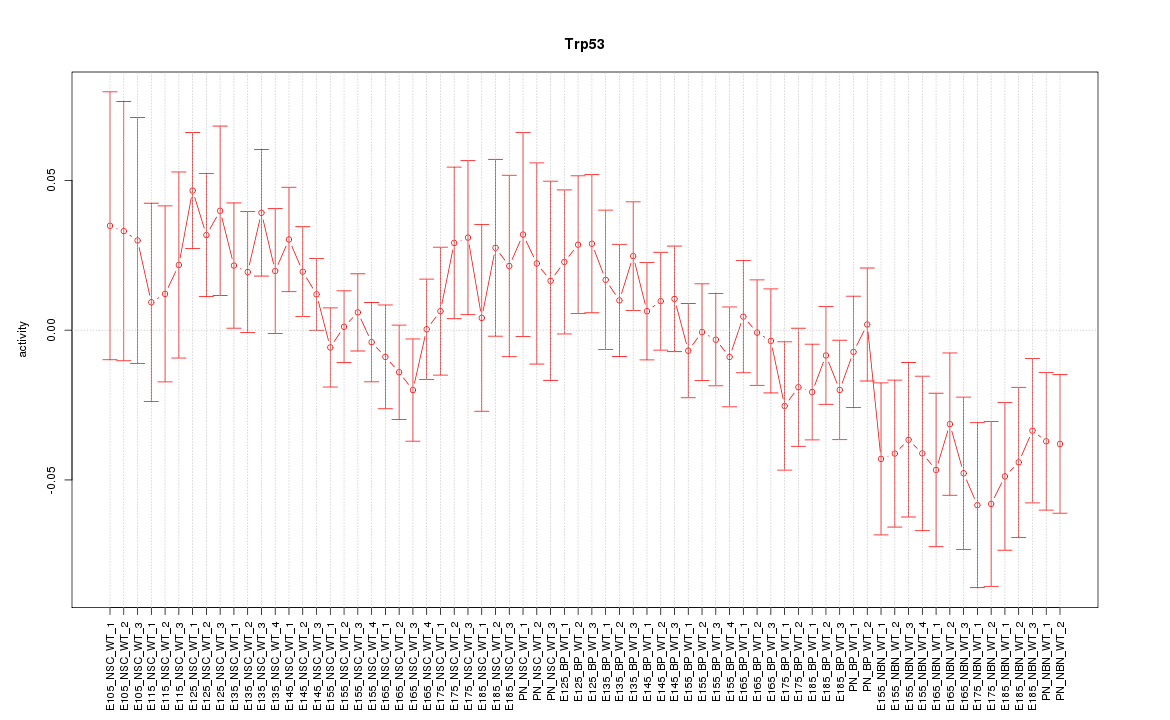
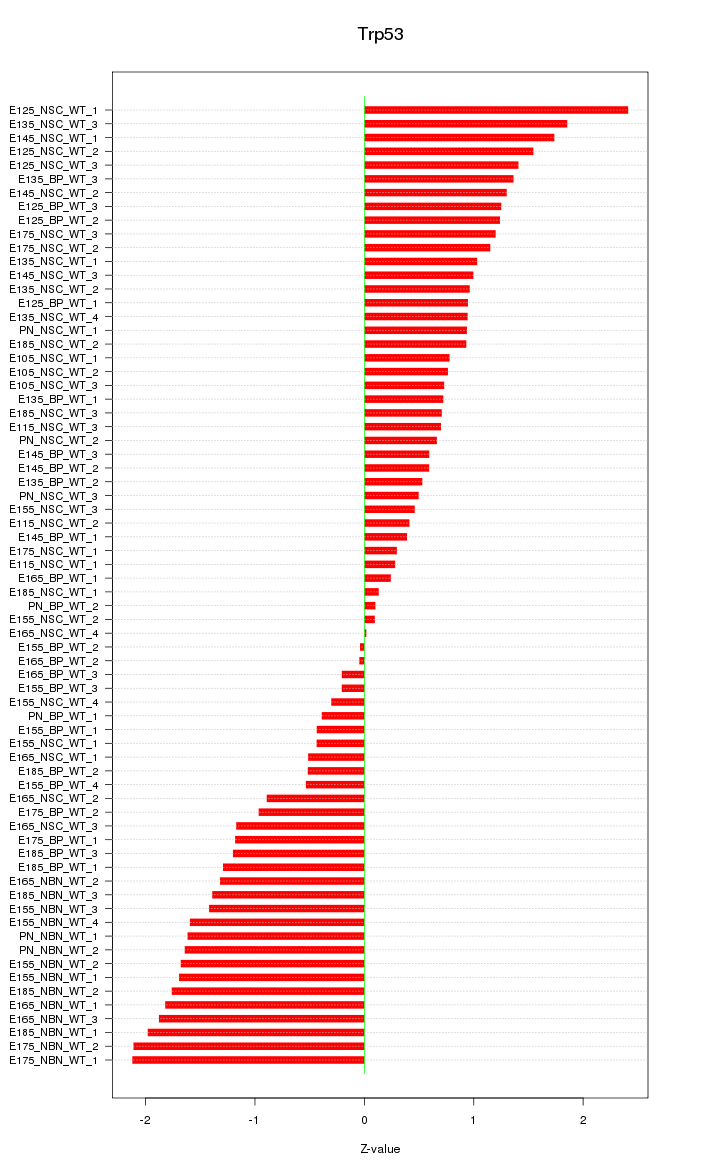
Motif ID: Trp53
Z-value: 1.131
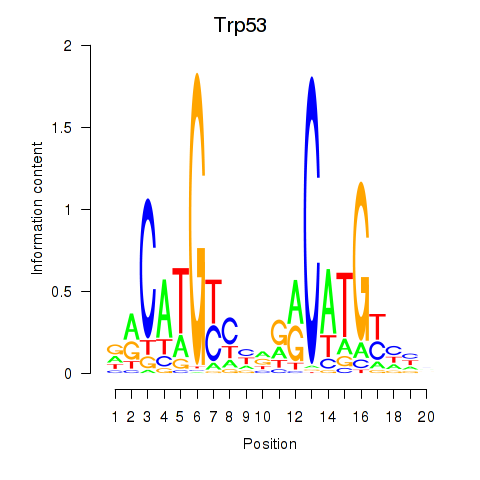
Transcription factors associated with Trp53:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Trp53 | ENSMUSG00000059552.7 | Trp53 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Trp53 | mm10_v2_chr11_+_69580392_69580419 | 0.49 | 1.7e-05 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.4 | 33.4 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 3.2 | 38.3 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 2.9 | 8.7 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 2.8 | 8.3 | GO:0003104 | positive regulation of glomerular filtration(GO:0003104) cell communication by chemical coupling(GO:0010643) |
| 2.4 | 7.3 | GO:0046078 | deoxyribonucleoside monophosphate biosynthetic process(GO:0009157) pyrimidine deoxyribonucleoside monophosphate biosynthetic process(GO:0009177) dUMP metabolic process(GO:0046078) |
| 2.3 | 7.0 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 1.6 | 4.7 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 1.5 | 4.5 | GO:0002765 | immune response-inhibiting signal transduction(GO:0002765) |
| 1.5 | 4.5 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 1.4 | 6.9 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 1.2 | 4.9 | GO:0099624 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) atrial cardiac muscle cell membrane repolarization(GO:0099624) |
| 1.1 | 3.2 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.9 | 2.7 | GO:1990046 | positive regulation of mitochondrial DNA replication(GO:0090297) regulation of cardiolipin metabolic process(GO:1900208) positive regulation of cardiolipin metabolic process(GO:1900210) stress-induced mitochondrial fusion(GO:1990046) |
| 0.9 | 13.3 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.9 | 6.8 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.8 | 12.8 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.8 | 2.3 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.7 | 2.1 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.7 | 4.3 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.7 | 3.4 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.6 | 2.4 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.6 | 4.6 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.5 | 4.7 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.5 | 2.0 | GO:0046909 | intermembrane transport(GO:0046909) |
| 0.4 | 1.8 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 0.4 | 1.3 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.4 | 1.7 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.4 | 7.1 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.4 | 34.8 | GO:0007052 | mitotic spindle organization(GO:0007052) |
| 0.4 | 1.1 | GO:0009812 | flavonoid metabolic process(GO:0009812) flavonoid biosynthetic process(GO:0009813) flavonoid glucuronidation(GO:0052696) |
| 0.4 | 23.2 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.3 | 2.0 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.3 | 3.4 | GO:0099623 | regulation of ventricular cardiac muscle cell membrane repolarization(GO:0060307) regulation of cardiac muscle cell membrane repolarization(GO:0099623) ventricular cardiac muscle cell membrane repolarization(GO:0099625) |
| 0.3 | 1.3 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.3 | 1.2 | GO:0006547 | histidine metabolic process(GO:0006547) |
| 0.3 | 2.6 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) sphingoid biosynthetic process(GO:0046520) |
| 0.2 | 4.2 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.2 | 5.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.2 | 1.9 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.2 | 2.1 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.2 | 1.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.2 | 1.5 | GO:1903204 | negative regulation of oxidative stress-induced neuron death(GO:1903204) |
| 0.2 | 1.2 | GO:0010694 | positive regulation of alkaline phosphatase activity(GO:0010694) |
| 0.2 | 4.6 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.2 | 2.5 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.2 | 2.7 | GO:0035337 | fatty-acyl-CoA metabolic process(GO:0035337) |
| 0.2 | 1.1 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.1 | 0.7 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.1 | 5.2 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 0.6 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.1 | 5.3 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.1 | 0.7 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.1 | 0.6 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.1 | 1.1 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 | 2.3 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.1 | 6.7 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.0 | 0.6 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 0.7 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.3 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.0 | 4.8 | GO:0009566 | fertilization(GO:0009566) |
| 0.0 | 0.8 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 1.6 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.3 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.2 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.4 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.0 | 2.9 | GO:0098780 | mitophagy in response to mitochondrial depolarization(GO:0098779) response to mitochondrial depolarisation(GO:0098780) |
| 0.0 | 0.9 | GO:0032611 | interleukin-1 beta production(GO:0032611) |
| 0.0 | 4.2 | GO:0009101 | glycoprotein biosynthetic process(GO:0009101) |
| 0.0 | 0.5 | GO:0006220 | pyrimidine nucleotide metabolic process(GO:0006220) |
| 0.0 | 2.6 | GO:0007018 | microtubule-based movement(GO:0007018) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.8 | 34.8 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 3.3 | 13.3 | GO:0002111 | BRCA2-BRAF35 complex(GO:0002111) |
| 2.6 | 33.9 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 2.3 | 6.8 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 2.1 | 33.4 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 1.7 | 10.1 | GO:0000235 | astral microtubule(GO:0000235) |
| 1.6 | 4.7 | GO:0033193 | Lsd1/2 complex(GO:0033193) |
| 0.9 | 9.0 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.8 | 8.7 | GO:0031091 | platelet alpha granule(GO:0031091) extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.8 | 8.3 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.6 | 3.4 | GO:0045179 | apical cortex(GO:0045179) |
| 0.5 | 2.3 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.4 | 4.5 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.4 | 23.5 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.2 | 1.2 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.2 | 1.3 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 0.2 | 3.4 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.2 | 2.7 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.2 | 2.5 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.2 | 10.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.2 | 2.1 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) cytosolic proteasome complex(GO:0031597) |
| 0.2 | 0.6 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.1 | 1.1 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.1 | 1.6 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 5.3 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 1.7 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 5.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 1.3 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 6.7 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 10.3 | GO:0043679 | axon terminus(GO:0043679) |
| 0.1 | 4.7 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 2.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.4 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.6 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 8.0 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.4 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.7 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.5 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 3.3 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 8.1 | GO:0019866 | mitochondrial inner membrane(GO:0005743) organelle inner membrane(GO:0019866) |
| 0.0 | 1.9 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.4 | 33.4 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 2.3 | 7.0 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 2.3 | 33.9 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 1.7 | 8.3 | GO:0071253 | connexin binding(GO:0071253) |
| 1.5 | 4.5 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 1.1 | 4.5 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 1.0 | 6.9 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 1.0 | 5.9 | GO:0008494 | translation activator activity(GO:0008494) |
| 1.0 | 4.9 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.9 | 2.7 | GO:1901611 | phosphatidylglycerol binding(GO:1901611) cardiolipin binding(GO:1901612) |
| 0.8 | 4.2 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.7 | 10.1 | GO:1990939 | ATP-dependent microtubule motor activity(GO:1990939) |
| 0.7 | 4.6 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.6 | 13.3 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.6 | 1.8 | GO:0018738 | S-formylglutathione hydrolase activity(GO:0018738) |
| 0.6 | 1.7 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.5 | 6.8 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.5 | 3.4 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.4 | 7.3 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.4 | 2.6 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.4 | 1.2 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.4 | 1.5 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.3 | 7.5 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.3 | 4.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.2 | 1.9 | GO:0046790 | virion binding(GO:0046790) |
| 0.2 | 0.6 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.2 | 8.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 1.6 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.2 | 2.5 | GO:0016594 | glycine binding(GO:0016594) |
| 0.2 | 1.5 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 5.2 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 4.6 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 0.7 | GO:0034916 | 4-methyloctanoyl-CoA dehydrogenase activity(GO:0034580) naphthyl-2-methyl-succinyl-CoA dehydrogenase activity(GO:0034845) 2-methylhexanoyl-CoA dehydrogenase activity(GO:0034916) propionyl-CoA dehydrogenase activity(GO:0043820) thiol-driven fumarate reductase activity(GO:0043830) coenzyme F420-dependent 2,4,6-trinitrophenol reductase activity(GO:0052758) coenzyme F420-dependent 2,4,6-trinitrophenol hydride reductase activity(GO:0052759) coenzyme F420-dependent 2,4-dinitrophenol reductase activity(GO:0052760) |
| 0.1 | 0.5 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.1 | 1.3 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.1 | 5.8 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 1.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.1 | 4.2 | GO:0050694 | galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 1.1 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.1 | 0.3 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 4.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 2.1 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 5.6 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 1.8 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.1 | 5.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 5.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.9 | GO:0052890 | acyl-CoA dehydrogenase activity(GO:0003995) oxidoreductase activity, acting on the CH-CH group of donors, with a flavin as acceptor(GO:0052890) |
| 0.1 | 6.0 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 0.4 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 2.3 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 2.9 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.5 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 2.7 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.7 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 3.2 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 0.2 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.6 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 1.1 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 1.2 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.0 | GO:0051219 | phosphoprotein binding(GO:0051219) |