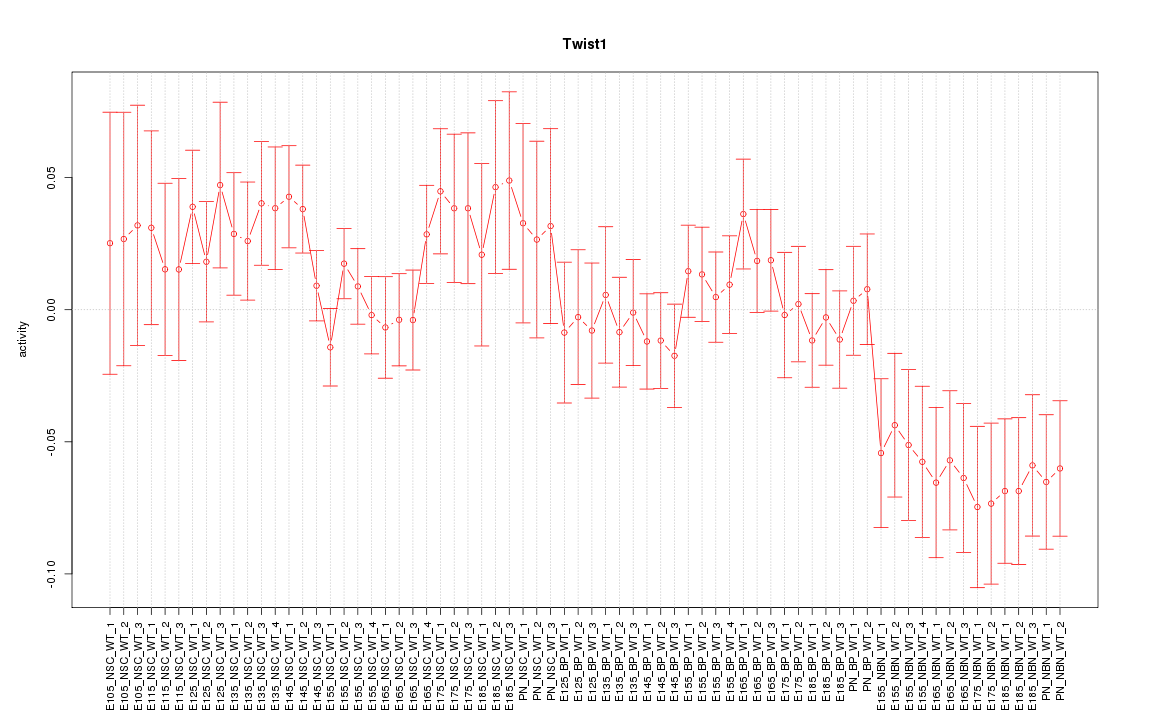
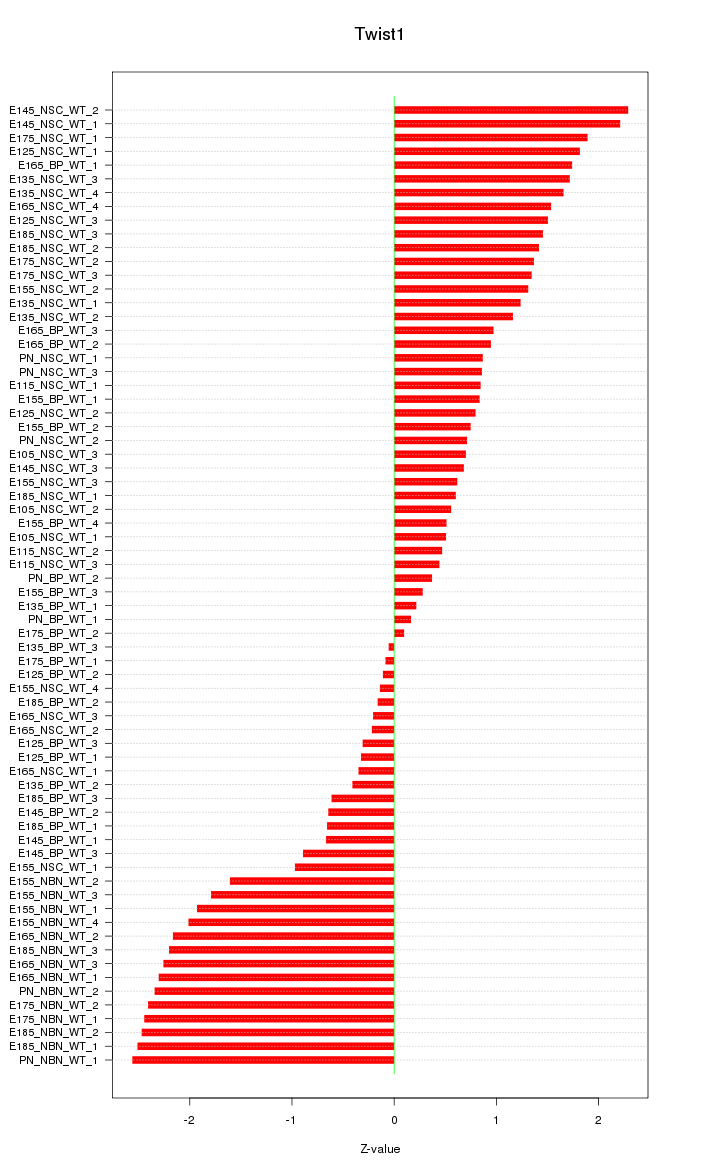
Motif ID: Twist1
Z-value: 1.343
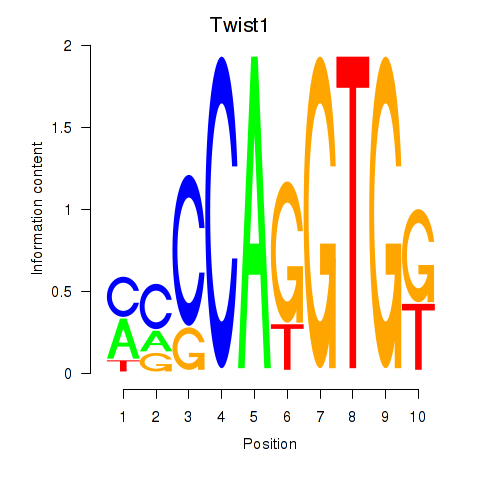
Transcription factors associated with Twist1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Twist1 | ENSMUSG00000035799.5 | Twist1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Twist1 | mm10_v2_chr12_+_33957645_33957671 | 0.15 | 2.1e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.7 | 34.7 | GO:0060032 | notochord regression(GO:0060032) |
| 6.1 | 6.1 | GO:1902460 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 5.9 | 17.7 | GO:0072076 | hyaluranon cable assembly(GO:0036118) nephrogenic mesenchyme development(GO:0072076) negative regulation of glomerular mesangial cell proliferation(GO:0072125) negative regulation of glomerulus development(GO:0090194) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 5.0 | 15.0 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 3.8 | 15.2 | GO:0051311 | spindle assembly involved in female meiosis(GO:0007056) meiotic metaphase plate congression(GO:0051311) |
| 2.9 | 8.8 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 2.2 | 6.5 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) dermatome development(GO:0061054) regulation of dermatome development(GO:0061183) negative regulation of dopaminergic neuron differentiation(GO:1904339) regulation of cardiac cell fate specification(GO:2000043) |
| 1.9 | 9.5 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 1.9 | 5.6 | GO:1904395 | regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904393) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 1.7 | 15.7 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 1.7 | 8.4 | GO:0072015 | ciliary body morphogenesis(GO:0061073) glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 1.6 | 14.5 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 1.2 | 3.6 | GO:2000564 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) CD8-positive, alpha-beta T cell proliferation(GO:0035740) negative regulation of regulatory T cell differentiation(GO:0045590) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 1.1 | 3.4 | GO:0019405 | alditol catabolic process(GO:0019405) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 1.0 | 3.1 | GO:0038145 | interleukin-4-mediated signaling pathway(GO:0035771) macrophage colony-stimulating factor signaling pathway(GO:0038145) tyrosine phosphorylation of Stat6 protein(GO:0042505) regulation of tyrosine phosphorylation of Stat6 protein(GO:0042525) regulation of interleukin-6-mediated signaling pathway(GO:0070103) negative regulation of interleukin-6-mediated signaling pathway(GO:0070104) |
| 1.0 | 2.9 | GO:0030070 | insulin processing(GO:0030070) |
| 1.0 | 9.5 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.9 | 17.1 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.7 | 13.4 | GO:0030903 | notochord development(GO:0030903) |
| 0.7 | 24.5 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.7 | 6.6 | GO:0018065 | protein-cofactor linkage(GO:0018065) |
| 0.7 | 2.0 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.6 | 4.5 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.6 | 6.2 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.5 | 2.2 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 0.5 | 18.5 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.5 | 2.6 | GO:0060838 | lymphatic endothelial cell fate commitment(GO:0060838) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.5 | 3.0 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.5 | 5.9 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.4 | 2.2 | GO:0019230 | protein autoprocessing(GO:0016540) proprioception(GO:0019230) |
| 0.4 | 3.0 | GO:0051503 | adenine nucleotide transport(GO:0051503) |
| 0.4 | 4.1 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.4 | 1.2 | GO:0060023 | soft palate development(GO:0060023) |
| 0.4 | 3.8 | GO:0050965 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.4 | 0.4 | GO:0061349 | planar cell polarity pathway involved in outflow tract morphogenesis(GO:0061347) planar cell polarity pathway involved in ventricular septum morphogenesis(GO:0061348) planar cell polarity pathway involved in cardiac right atrium morphogenesis(GO:0061349) planar cell polarity pathway involved in cardiac muscle tissue morphogenesis(GO:0061350) planar cell polarity pathway involved in pericardium morphogenesis(GO:0061354) |
| 0.3 | 2.8 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.3 | 12.7 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.2 | 3.9 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.2 | 0.9 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.2 | 1.1 | GO:0032911 | nerve growth factor production(GO:0032902) negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.2 | 1.7 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 4.5 | GO:0030201 | heparan sulfate proteoglycan metabolic process(GO:0030201) |
| 0.1 | 2.7 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 3.2 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.1 | 1.3 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.1 | 0.5 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 | 0.9 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 0.4 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.1 | 0.6 | GO:0033014 | porphyrin-containing compound biosynthetic process(GO:0006779) tetrapyrrole biosynthetic process(GO:0033014) |
| 0.1 | 11.3 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.1 | 0.9 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.1 | 0.6 | GO:0060766 | negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.0 | 1.6 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.6 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 1.0 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 3.7 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.0 | 3.5 | GO:0042594 | response to starvation(GO:0042594) |
| 0.0 | 4.7 | GO:0006909 | phagocytosis(GO:0006909) |
| 0.0 | 1.1 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 4.4 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.0 | 0.7 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 2.5 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.3 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 | 0.4 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.4 | GO:0030488 | tRNA methylation(GO:0030488) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.9 | 17.6 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 4.3 | 17.1 | GO:0043293 | apoptosome(GO:0043293) |
| 3.5 | 14.0 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 2.9 | 8.8 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 2.7 | 34.7 | GO:0097542 | ciliary tip(GO:0097542) |
| 2.4 | 14.5 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 2.3 | 6.8 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 1.7 | 15.2 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.9 | 3.7 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.7 | 5.9 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.6 | 1.8 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.2 | 6.2 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.2 | 5.3 | GO:0002102 | podosome(GO:0002102) |
| 0.2 | 2.9 | GO:0031045 | dense core granule(GO:0031045) |
| 0.2 | 0.9 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 24.5 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 2.2 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.1 | 1.2 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 28.8 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 1.0 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 1.1 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.1 | 8.6 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 1.6 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 0.4 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 3.0 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 0.7 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 3.1 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 6.6 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 11.3 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 4.1 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 2.0 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.2 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.6 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 1.2 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 0.5 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 10.2 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 3.6 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 2.0 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.4 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 2.1 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 15.3 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.4 | 17.6 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 2.4 | 14.5 | GO:0034943 | acyl-CoA ligase activity(GO:0003996) 3-oxo-2-(2'-pentenyl)cyclopentane-1-octanoic acid CoA ligase activity(GO:0010435) 3-isopropenyl-6-oxoheptanoyl-CoA synthetase activity(GO:0018854) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA synthetase activity(GO:0018855) benzoyl acetate-CoA ligase activity(GO:0018856) 2,4-dichlorobenzoate-CoA ligase activity(GO:0018857) pivalate-CoA ligase activity(GO:0034783) cyclopropanecarboxylate-CoA ligase activity(GO:0034793) adipate-CoA ligase activity(GO:0034796) citronellyl-CoA ligase activity(GO:0034823) mentha-1,3-dione-CoA ligase activity(GO:0034841) thiophene-2-carboxylate-CoA ligase activity(GO:0034842) 2,4,4-trimethylpentanoate-CoA ligase activity(GO:0034865) cis-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034942) trans-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034943) branched-chain acyl-CoA synthetase (ADP-forming) activity(GO:0043759) aryl-CoA synthetase (ADP-forming) activity(GO:0043762) 3-hydroxypropionyl-CoA synthetase activity(GO:0043955) perillic acid:CoA ligase (ADP-forming) activity(GO:0052685) perillic acid:CoA ligase (AMP-forming) activity(GO:0052686) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (ADP-forming) activity(GO:0052687) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (AMP-forming) activity(GO:0052688) pristanate-CoA ligase activity(GO:0070251) malonyl-CoA synthetase activity(GO:0090409) |
| 2.2 | 24.5 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 2.2 | 6.5 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 1.8 | 17.7 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 1.3 | 10.5 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 1.1 | 8.8 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 1.1 | 6.6 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 1.0 | 3.0 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 1.0 | 9.5 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.7 | 7.4 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.7 | 10.1 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.7 | 4.1 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.4 | 3.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.4 | 2.2 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.3 | 6.0 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.3 | 9.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.3 | 4.5 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.3 | 3.4 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.3 | 2.6 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.3 | 34.1 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.3 | 8.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.3 | 6.1 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.3 | 15.0 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.2 | 2.9 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) cobalt ion binding(GO:0050897) |
| 0.2 | 2.0 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.2 | 1.6 | GO:0016668 | oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor(GO:0016668) |
| 0.2 | 6.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.2 | 6.2 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.1 | 0.4 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.1 | 6.7 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 14.0 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 1.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 4.4 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.1 | 2.2 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.1 | 2.2 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 3.8 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.1 | 29.0 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.1 | 0.6 | GO:0034916 | 4-methyloctanoyl-CoA dehydrogenase activity(GO:0034580) naphthyl-2-methyl-succinyl-CoA dehydrogenase activity(GO:0034845) 2-methylhexanoyl-CoA dehydrogenase activity(GO:0034916) propionyl-CoA dehydrogenase activity(GO:0043820) thiol-driven fumarate reductase activity(GO:0043830) coenzyme F420-dependent 2,4,6-trinitrophenol reductase activity(GO:0052758) coenzyme F420-dependent 2,4,6-trinitrophenol hydride reductase activity(GO:0052759) coenzyme F420-dependent 2,4-dinitrophenol reductase activity(GO:0052760) |
| 0.1 | 0.6 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 2.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 6.1 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.1 | 0.5 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.1 | 0.7 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 1.7 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 9.2 | GO:0000987 | core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.1 | 5.3 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 1.1 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.0 | 15.2 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 1.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.4 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.9 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) |
| 0.0 | 1.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 4.1 | GO:0008757 | S-adenosylmethionine-dependent methyltransferase activity(GO:0008757) |
| 0.0 | 3.5 | GO:0003729 | mRNA binding(GO:0003729) |
| 0.0 | 3.2 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.7 | GO:0002039 | p53 binding(GO:0002039) |