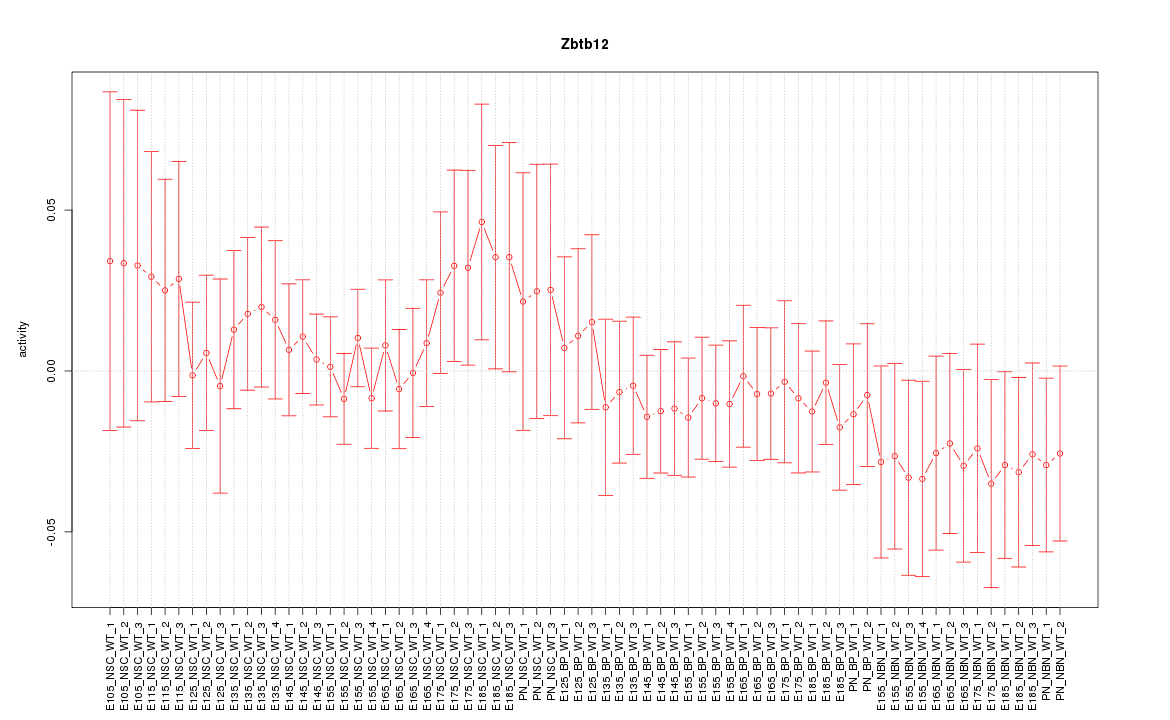
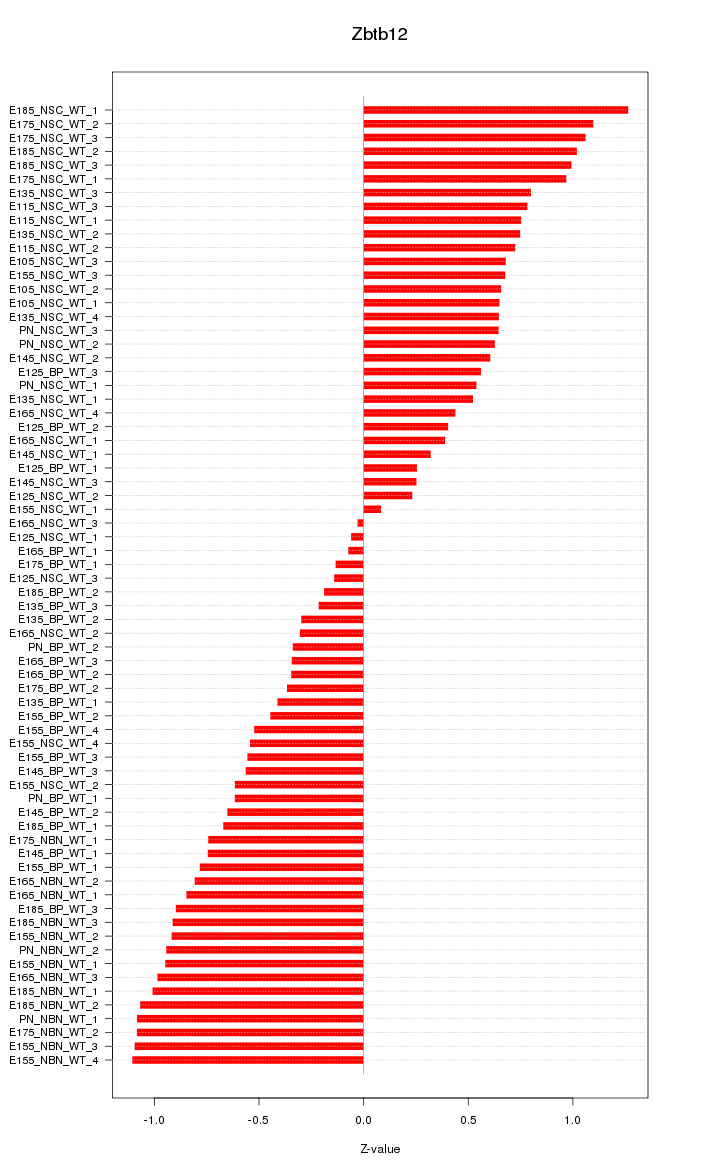
Motif ID: Zbtb12
Z-value: 0.698
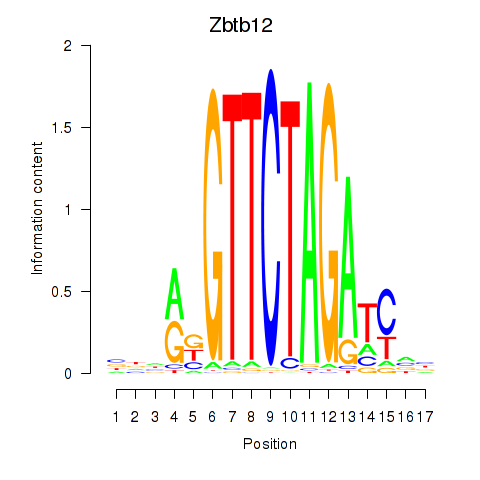
Transcription factors associated with Zbtb12:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Zbtb12 | ENSMUSG00000049823.8 | Zbtb12 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb12 | mm10_v2_chr17_+_34894515_34894573 | -0.42 | 3.5e-04 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 9.8 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 2.3 | 6.9 | GO:0019405 | alditol catabolic process(GO:0019405) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 1.8 | 5.4 | GO:0060853 | arterial endothelial cell fate commitment(GO:0060844) blood vessel endothelial cell fate commitment(GO:0060846) endothelial cell fate specification(GO:0060847) Notch signaling pathway involved in arterial endothelial cell fate commitment(GO:0060853) blood vessel endothelial cell fate specification(GO:0097101) |
| 1.7 | 5.2 | GO:0010248 | B cell negative selection(GO:0002352) establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 1.3 | 11.8 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 1.1 | 6.3 | GO:0098700 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 1.0 | 8.2 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 1.0 | 2.9 | GO:1900275 | negative regulation of phospholipase C activity(GO:1900275) |
| 0.8 | 2.4 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.7 | 2.1 | GO:0002586 | positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.5 | 1.4 | GO:0009174 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.4 | 9.7 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.4 | 1.9 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.4 | 5.7 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.3 | 1.0 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.3 | 3.0 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.3 | 4.4 | GO:0072010 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.3 | 2.3 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) regulation of thyroid hormone mediated signaling pathway(GO:0002155) |
| 0.2 | 0.6 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.2 | 0.5 | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.1 | 2.4 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 | 2.9 | GO:0005980 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.1 | 0.4 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.1 | 3.0 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.3 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 8.2 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 2.5 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 0.6 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 1.7 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.2 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) response to glucagon(GO:0033762) |
| 0.0 | 6.1 | GO:0051321 | meiotic cell cycle(GO:0051321) |
| 0.0 | 4.9 | GO:0048864 | stem cell development(GO:0048864) |
| 0.0 | 0.3 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.2 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.0 | 0.4 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.4 | GO:0031440 | regulation of mRNA 3'-end processing(GO:0031440) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.0 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.5 | 2.1 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.4 | 6.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.4 | 5.2 | GO:0046930 | pore complex(GO:0046930) |
| 0.2 | 5.7 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.1 | 1.9 | GO:0044447 | axoneme part(GO:0044447) |
| 0.1 | 8.0 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 4.9 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.6 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 9.8 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 2.3 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 0.3 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.5 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 1.7 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 0.6 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 5.4 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 3.3 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 2.9 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 18.2 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.2 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 8.2 | GO:0030424 | axon(GO:0030424) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 9.8 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 2.1 | 6.3 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 1.6 | 8.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.7 | 2.9 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.6 | 9.7 | GO:0008430 | selenium binding(GO:0008430) |
| 0.6 | 6.9 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.5 | 11.8 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.5 | 5.4 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.4 | 2.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.4 | 5.2 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.3 | 1.0 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.3 | 2.1 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.2 | 2.9 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.2 | 5.7 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.2 | 4.9 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 0.3 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 0.4 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.3 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.6 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 1.4 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.5 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 3.0 | GO:0008234 | cysteine-type peptidase activity(GO:0008234) |
| 0.0 | 4.8 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.2 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 2.6 | GO:0003924 | GTPase activity(GO:0003924) |