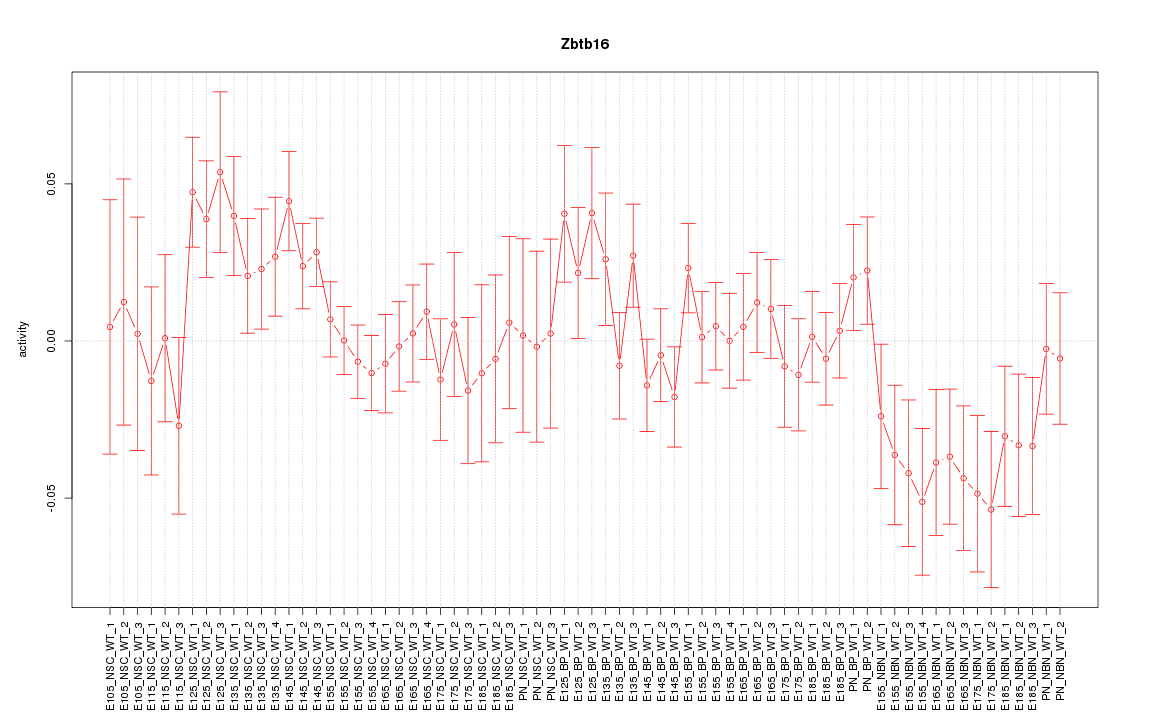
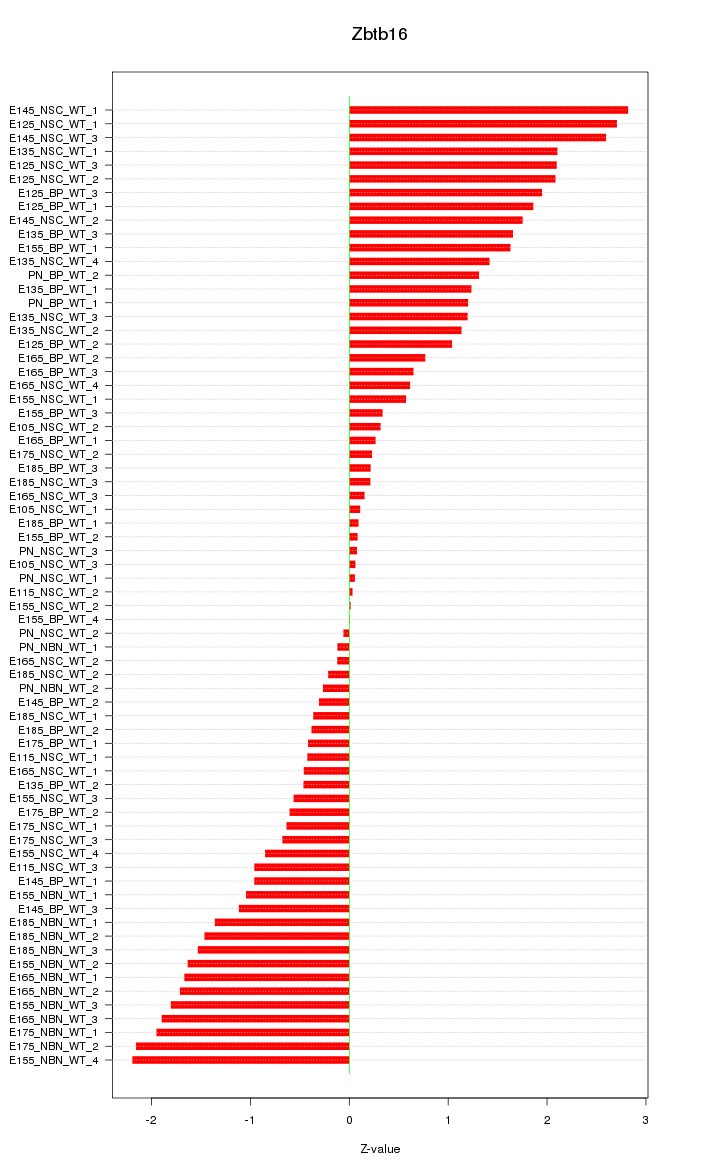
Motif ID: Zbtb16
Z-value: 1.232
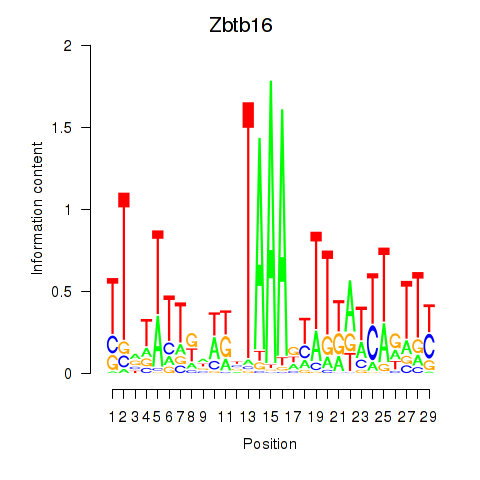
Transcription factors associated with Zbtb16:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Zbtb16 | ENSMUSG00000066687.4 | Zbtb16 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb16 | mm10_v2_chr9_-_48835932_48835962 | -0.07 | 5.4e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 11.0 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 2.9 | 14.5 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 2.7 | 8.0 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 1.7 | 7.0 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 1.7 | 8.3 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 1.6 | 4.9 | GO:1905065 | cysteine biosynthetic process via cystathionine(GO:0019343) cysteine biosynthetic process(GO:0019344) positive regulation of vascular smooth muscle cell differentiation(GO:1905065) |
| 0.9 | 3.5 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.8 | 3.3 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.8 | 2.4 | GO:0045819 | positive regulation of glycogen catabolic process(GO:0045819) |
| 0.8 | 4.5 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.7 | 7.4 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.4 | 1.7 | GO:0032382 | positive regulation of intracellular lipid transport(GO:0032379) positive regulation of intracellular sterol transport(GO:0032382) positive regulation of intracellular cholesterol transport(GO:0032385) lipid hydroperoxide transport(GO:1901373) positive regulation of cholesterol import(GO:1904109) positive regulation of sterol import(GO:2000911) |
| 0.4 | 2.6 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.4 | 2.1 | GO:0036337 | Fas signaling pathway(GO:0036337) |
| 0.4 | 1.3 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.4 | 1.5 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.4 | 1.9 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.4 | 1.4 | GO:0006203 | dGTP catabolic process(GO:0006203) |
| 0.3 | 1.4 | GO:0072592 | regulation of integrin biosynthetic process(GO:0045113) oxygen metabolic process(GO:0072592) |
| 0.3 | 19.2 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) |
| 0.3 | 0.9 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.3 | 2.0 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.3 | 1.6 | GO:0051461 | regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.3 | 14.9 | GO:0008631 | intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0008631) |
| 0.2 | 3.6 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.2 | 1.4 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.2 | 6.4 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.2 | 1.7 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.2 | 1.5 | GO:0019370 | leukotriene biosynthetic process(GO:0019370) |
| 0.2 | 0.6 | GO:1902524 | positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 0.2 | 0.6 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.2 | 4.0 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.2 | 1.2 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.2 | 2.3 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.2 | 1.2 | GO:0043144 | snoRNA processing(GO:0043144) |
| 0.1 | 0.9 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.1 | 1.4 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.1 | 3.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 0.9 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.1 | 1.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.4 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.1 | 0.9 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.1 | 0.6 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 1.7 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.1 | 0.5 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 | 0.6 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.1 | 4.6 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.1 | 0.5 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.1 | 2.1 | GO:0001913 | T cell mediated cytotoxicity(GO:0001913) |
| 0.1 | 1.1 | GO:0000479 | endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.1 | 0.8 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.1 | 11.9 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.1 | 0.4 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.1 | 0.6 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.1 | 0.5 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.1 | 0.7 | GO:0032460 | negative regulation of protein oligomerization(GO:0032460) negative regulation of protein homooligomerization(GO:0032463) |
| 0.1 | 2.5 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 3.8 | GO:0007569 | cell aging(GO:0007569) |
| 0.1 | 0.5 | GO:0006183 | GTP biosynthetic process(GO:0006183) |
| 0.1 | 1.2 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.1 | 0.5 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.1 | 0.4 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 3.3 | GO:0002066 | columnar/cuboidal epithelial cell development(GO:0002066) |
| 0.0 | 0.3 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.2 | GO:0048318 | axial mesoderm development(GO:0048318) |
| 0.0 | 2.4 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.4 | GO:0030539 | male genitalia development(GO:0030539) |
| 0.0 | 0.5 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 1.1 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.0 | 0.2 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 3.7 | GO:0042472 | inner ear morphogenesis(GO:0042472) |
| 0.0 | 1.6 | GO:0072661 | protein targeting to plasma membrane(GO:0072661) |
| 0.0 | 1.7 | GO:0045843 | negative regulation of striated muscle tissue development(GO:0045843) |
| 0.0 | 3.1 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 1.4 | GO:0009214 | cyclic nucleotide catabolic process(GO:0009214) |
| 0.0 | 0.2 | GO:0071267 | amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.0 | 0.2 | GO:0051189 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 2.2 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.4 | GO:0010388 | cullin deneddylation(GO:0010388) |
| 0.0 | 0.7 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.0 | 2.2 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.9 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 0.2 | GO:0015838 | amino-acid betaine transport(GO:0015838) |
| 0.0 | 0.4 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.0 | 2.3 | GO:0042493 | response to drug(GO:0042493) |
| 0.0 | 0.6 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.1 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) positive regulation of extracellular exosome assembly(GO:1903553) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 11.0 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 1.4 | 8.3 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.9 | 4.5 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.8 | 2.4 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.6 | 1.8 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.5 | 2.0 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.4 | 14.5 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.4 | 2.6 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.4 | 1.6 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.3 | 1.4 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.3 | 1.9 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.3 | 0.9 | GO:0030690 | Noc1p-Noc2p complex(GO:0030690) |
| 0.2 | 1.7 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.2 | 1.5 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.2 | 1.4 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.2 | 2.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.2 | 1.1 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 1.4 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.1 | 1.1 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 1.7 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.1 | 1.6 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.1 | 0.5 | GO:0001651 | dense fibrillar component(GO:0001651) granular component(GO:0001652) |
| 0.1 | 0.4 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 1.7 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 0.6 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 1.5 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 7.7 | GO:1990204 | oxidoreductase complex(GO:1990204) |
| 0.1 | 0.9 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.1 | 18.0 | GO:0005938 | cell cortex(GO:0005938) |
| 0.1 | 5.1 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 0.5 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 0.4 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 1.9 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.6 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.7 | GO:0005684 | U2-type spliceosomal complex(GO:0005684) |
| 0.0 | 0.7 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.6 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 3.8 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.6 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 3.3 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 0.4 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 1.2 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.6 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.2 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 7.3 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.6 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.4 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.9 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.8 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 8.3 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 1.6 | 11.0 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 1.0 | 14.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.8 | 2.4 | GO:0000402 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.7 | 2.2 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.7 | 3.6 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.7 | 8.5 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.6 | 4.9 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.5 | 4.6 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.5 | 8.0 | GO:0016594 | glycine binding(GO:0016594) |
| 0.5 | 1.4 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.4 | 1.7 | GO:0033814 | propanoyl-CoA C-acyltransferase activity(GO:0033814) propionyl-CoA C2-trimethyltridecanoyltransferase activity(GO:0050632) phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.4 | 1.7 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.4 | 1.3 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 0.4 | 1.2 | GO:0008534 | oxidized purine nucleobase lesion DNA N-glycosylase activity(GO:0008534) |
| 0.4 | 1.6 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.4 | 14.9 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.3 | 4.0 | GO:0034951 | pivalyl-CoA mutase activity(GO:0034784) o-hydroxylaminobenzoate mutase activity(GO:0034951) lupeol synthase activity(GO:0042299) beta-amyrin synthase activity(GO:0042300) baruol synthase activity(GO:0080011) |
| 0.3 | 1.6 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.3 | 2.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.3 | 2.0 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.3 | 2.1 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.3 | 11.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 2.6 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.2 | 2.6 | GO:0070636 | single-strand selective uracil DNA N-glycosylase activity(GO:0017065) nicotinamide riboside hydrolase activity(GO:0070635) nicotinic acid riboside hydrolase activity(GO:0070636) deoxyribonucleoside 5'-monophosphate N-glycosidase activity(GO:0070694) |
| 0.2 | 0.9 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.2 | 1.1 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.2 | 0.6 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.2 | 2.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.2 | 3.3 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.2 | 1.7 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.2 | 2.7 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 4.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 3.6 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 1.5 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.1 | 6.4 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.1 | 1.5 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.1 | 2.5 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 1.4 | GO:0008413 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) |
| 0.1 | 1.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 1.7 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 3.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 1.5 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 0.5 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.1 | 0.4 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.1 | 0.5 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.1 | 1.9 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 2.8 | GO:0008186 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 0.5 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.5 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 1.8 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.6 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.8 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.2 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 1.4 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 1.4 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.7 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 1.3 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 1.8 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 1.4 | GO:0030551 | cyclic nucleotide binding(GO:0030551) |
| 0.0 | 1.6 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.0 | 1.1 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 2.6 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 2.3 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.6 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 1.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 6.4 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.5 | GO:0004177 | aminopeptidase activity(GO:0004177) |