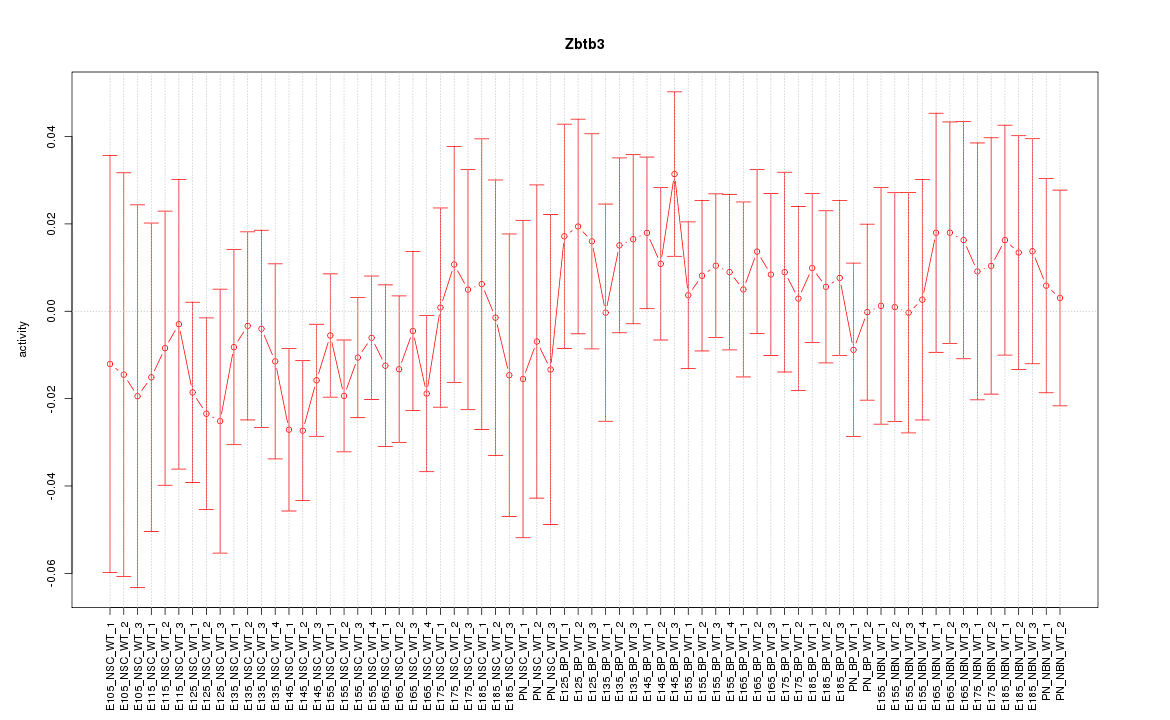
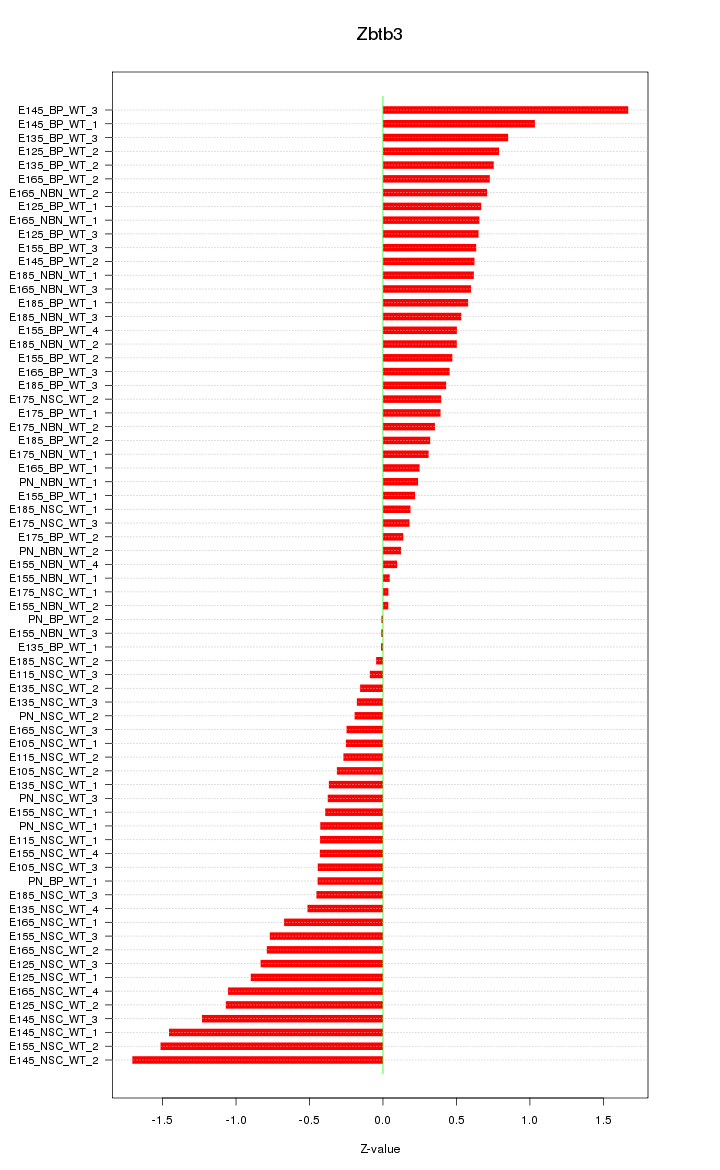
Motif ID: Zbtb3
Z-value: 0.642
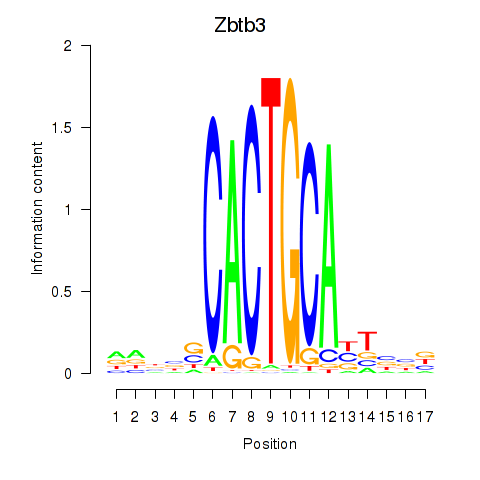
Transcription factors associated with Zbtb3:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Zbtb3 | ENSMUSG00000071661.6 | Zbtb3 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb3 | mm10_v2_chr19_+_8802486_8802530 | 0.49 | 1.7e-05 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 1.1 | GO:1900275 | negative regulation of phospholipase C activity(GO:1900275) |
| 0.9 | 2.8 | GO:0001978 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) |
| 0.6 | 2.2 | GO:1904529 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 0.4 | 3.6 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.3 | 1.0 | GO:0010512 | negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) |
| 0.3 | 3.4 | GO:0090232 | positive regulation of spindle checkpoint(GO:0090232) |
| 0.3 | 1.3 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.3 | 0.8 | GO:0060854 | patterning of lymph vessels(GO:0060854) |
| 0.2 | 0.6 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.2 | 1.1 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.2 | 0.7 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.2 | 0.5 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) ceramide transport(GO:0035627) |
| 0.1 | 1.9 | GO:2000009 | negative regulation of protein localization to cell surface(GO:2000009) |
| 0.1 | 2.0 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.1 | 0.6 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.1 | 0.4 | GO:0051204 | protein insertion into mitochondrial membrane(GO:0051204) |
| 0.1 | 0.8 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.1 | 0.3 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.1 | 0.5 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.1 | 2.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 1.2 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 0.4 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.1 | 1.3 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) membrane hyperpolarization(GO:0060081) |
| 0.1 | 0.2 | GO:0009838 | abscission(GO:0009838) |
| 0.1 | 0.6 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.8 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.1 | 3.2 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 1.5 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 1.2 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.1 | 1.2 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.1 | 0.8 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.1 | 0.5 | GO:1901030 | positive regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901030) |
| 0.1 | 1.4 | GO:0048148 | behavioral response to cocaine(GO:0048148) |
| 0.0 | 0.2 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.7 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.4 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.0 | 4.4 | GO:0014065 | phosphatidylinositol 3-kinase signaling(GO:0014065) |
| 0.0 | 0.6 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.3 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.0 | 1.0 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.0 | 0.6 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 1.4 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 0.2 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.3 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.0 | 1.2 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.6 | GO:0030262 | cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.0 | 0.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.2 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.2 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 1.0 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.5 | GO:0060135 | maternal process involved in female pregnancy(GO:0060135) |
| 0.0 | 0.5 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.8 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.2 | 2.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.2 | 1.1 | GO:0033648 | host intracellular organelle(GO:0033647) host intracellular membrane-bounded organelle(GO:0033648) |
| 0.1 | 1.3 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 1.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 1.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 3.4 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 3.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 4.0 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 0.4 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 2.2 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 1.8 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.1 | 0.8 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.1 | 0.4 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 2.3 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.2 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 2.7 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 1.5 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.6 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.5 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.0 | 0.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 2.4 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.2 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.7 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.7 | GO:0005902 | microvillus(GO:0005902) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.4 | 1.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.3 | 3.6 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.3 | 2.8 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.3 | 2.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.3 | 1.5 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.2 | 1.4 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.2 | 0.5 | GO:0042030 | ATPase inhibitor activity(GO:0042030) angiostatin binding(GO:0043532) |
| 0.2 | 0.5 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.1 | 2.2 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 0.5 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.1 | 1.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.5 | GO:0097001 | sphingolipid transporter activity(GO:0046624) ceramide binding(GO:0097001) |
| 0.1 | 1.8 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 1.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.3 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.1 | 3.4 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 1.0 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.7 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.1 | 0.6 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.1 | 0.6 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.1 | 1.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 1.0 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.2 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.0 | 0.4 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.1 | GO:0001888 | glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0001888) |
| 0.0 | 1.8 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.6 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 4.2 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 1.3 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.3 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.4 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 1.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.4 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 3.6 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 2.7 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.8 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.5 | GO:0005246 | calcium channel regulator activity(GO:0005246) |