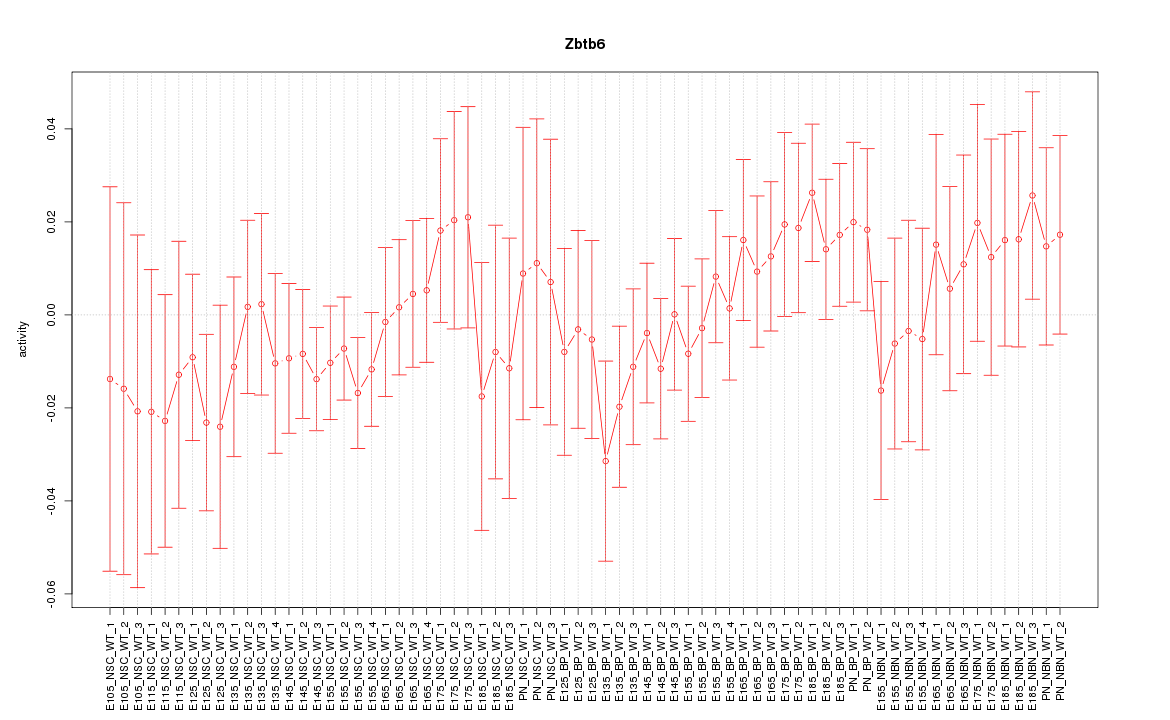
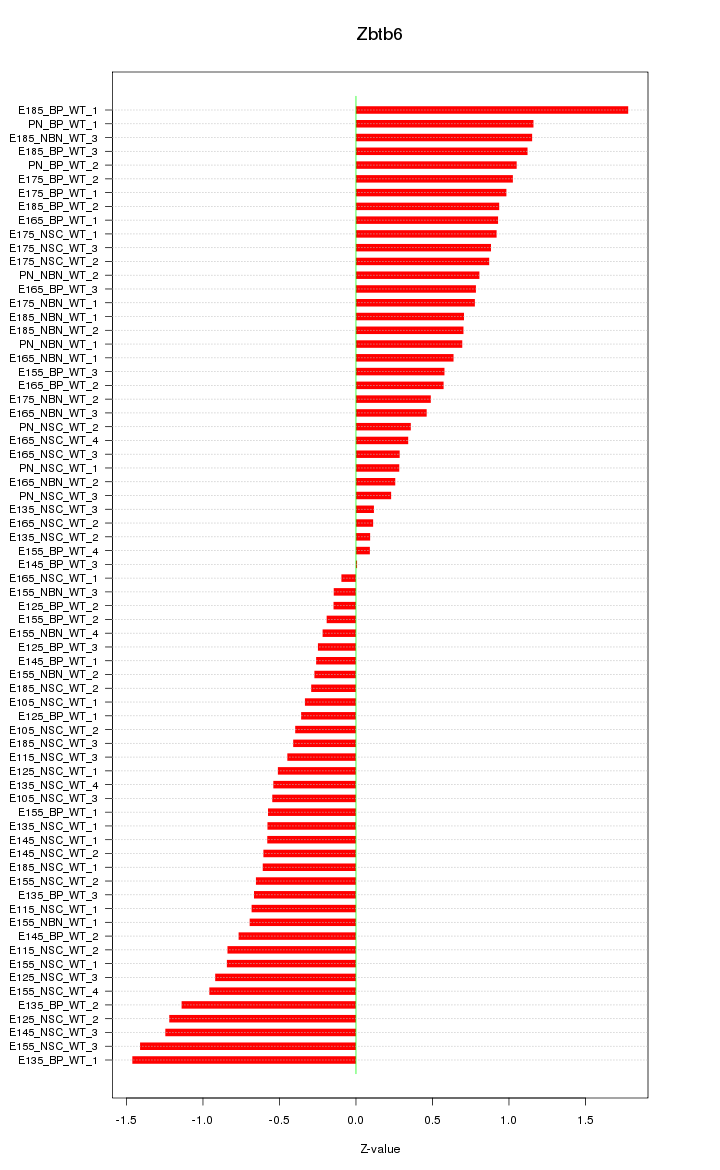
Motif ID: Zbtb6
Z-value: 0.733
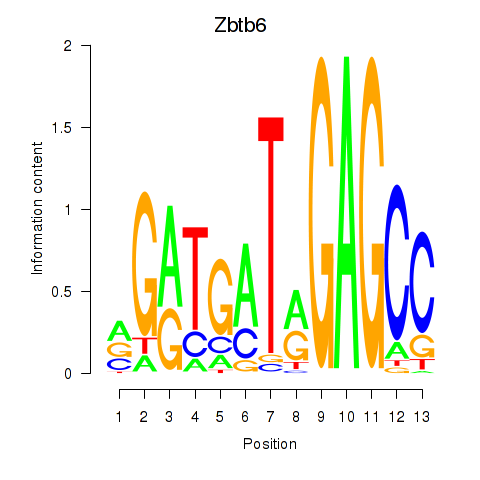
Transcription factors associated with Zbtb6:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Zbtb6 | ENSMUSG00000066798.3 | Zbtb6 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb6 | mm10_v2_chr2_-_37430835_37430919 | 0.45 | 8.9e-05 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 10.3 | GO:0010624 | regulation of Schwann cell proliferation(GO:0010624) |
| 1.3 | 3.8 | GO:0021837 | motogenic signaling involved in postnatal olfactory bulb interneuron migration(GO:0021837) positive regulation of mitotic cell cycle DNA replication(GO:1903465) |
| 1.3 | 5.0 | GO:0009414 | response to water deprivation(GO:0009414) |
| 1.0 | 3.0 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 0.6 | 1.9 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.6 | 2.3 | GO:0010040 | response to iron(II) ion(GO:0010040) positive regulation of hydrogen peroxide metabolic process(GO:0010726) negative regulation of synaptic transmission, dopaminergic(GO:0032227) negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) cellular response to copper ion(GO:0071280) regulation of peroxidase activity(GO:2000468) |
| 0.5 | 3.3 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.4 | 4.5 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.4 | 1.3 | GO:2000564 | CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) positive regulation of CD8-positive, alpha-beta T cell activation(GO:2001187) |
| 0.4 | 9.0 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.4 | 1.3 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.4 | 2.6 | GO:0036006 | response to macrophage colony-stimulating factor(GO:0036005) cellular response to macrophage colony-stimulating factor stimulus(GO:0036006) |
| 0.4 | 1.1 | GO:0030070 | insulin processing(GO:0030070) |
| 0.4 | 3.6 | GO:0007379 | segment specification(GO:0007379) |
| 0.4 | 1.1 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.3 | 5.6 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.3 | 1.0 | GO:0048388 | endosomal lumen acidification(GO:0048388) synaptic vesicle lumen acidification(GO:0097401) |
| 0.3 | 1.0 | GO:0030321 | transepithelial chloride transport(GO:0030321) transepithelial ammonium transport(GO:0070634) |
| 0.3 | 0.7 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.3 | 1.6 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.3 | 1.5 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.3 | 1.8 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.3 | 1.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.3 | 0.8 | GO:0050929 | corticospinal neuron axon guidance through spinal cord(GO:0021972) positive regulation of negative chemotaxis(GO:0050924) induction of negative chemotaxis(GO:0050929) negative regulation of mononuclear cell migration(GO:0071676) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.3 | 0.8 | GO:0061743 | motor learning(GO:0061743) |
| 0.3 | 1.3 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.3 | 1.8 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.2 | 2.2 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.2 | 0.5 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.2 | 2.5 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.2 | 0.6 | GO:0043201 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) |
| 0.2 | 1.5 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.2 | 1.9 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.2 | 1.2 | GO:0048840 | otolith development(GO:0048840) |
| 0.2 | 1.0 | GO:1901678 | iron coordination entity transport(GO:1901678) |
| 0.2 | 0.7 | GO:0060744 | thelarche(GO:0042695) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) mammary gland branching involved in thelarche(GO:0060744) mammary gland branching involved in pregnancy(GO:0060745) positive regulation of G0 to G1 transition(GO:0070318) |
| 0.1 | 4.6 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 2.0 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 | 1.0 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 0.5 | GO:0001907 | killing by symbiont of host cells(GO:0001907) induction of programmed cell death(GO:0012502) disruption by symbiont of host cell(GO:0044004) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.1 | 1.9 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 0.4 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.1 | 2.1 | GO:0021702 | cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.1 | 0.7 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 | 0.7 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 0.8 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.1 | 0.5 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 0.6 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 1.6 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.1 | 0.5 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.1 | 0.8 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.1 | 1.1 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 1.2 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.1 | 0.6 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 0.3 | GO:1903912 | negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:1903912) |
| 0.1 | 1.7 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 1.4 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.1 | GO:0060763 | mammary duct terminal end bud growth(GO:0060763) |
| 0.0 | 0.4 | GO:1900107 | regulation of nodal signaling pathway(GO:1900107) |
| 0.0 | 0.2 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.4 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.7 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 1.7 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 1.0 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 1.6 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 2.9 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.2 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 0.4 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.0 | 1.2 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 0.3 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.0 | 0.5 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 1.2 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.0 | 1.8 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.9 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.6 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.6 | GO:0009268 | response to pH(GO:0009268) |
| 0.0 | 0.7 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.4 | GO:0055012 | ventricular cardiac muscle cell differentiation(GO:0055012) |
| 0.0 | 0.9 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.0 | 0.1 | GO:0045629 | positive regulation of T-helper 1 cell differentiation(GO:0045627) negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.2 | GO:0061000 | negative regulation of dendritic spine development(GO:0061000) |
| 0.0 | 2.2 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.0 | 2.3 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.5 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 1.3 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 1.1 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.0 | GO:0032916 | positive regulation of transforming growth factor beta3 production(GO:0032916) |
| 0.0 | 0.7 | GO:0035773 | insulin secretion involved in cellular response to glucose stimulus(GO:0035773) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 10.3 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.6 | 2.3 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.4 | 1.7 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.4 | 1.2 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.2 | 0.7 | GO:0055087 | Ski complex(GO:0055087) |
| 0.2 | 0.8 | GO:0044301 | climbing fiber(GO:0044301) |
| 0.1 | 1.9 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 2.2 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 1.1 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 1.9 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.1 | 1.1 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 1.3 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 4.7 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.1 | 1.0 | GO:0042581 | specific granule(GO:0042581) |
| 0.1 | 2.7 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.1 | 0.8 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 0.6 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.1 | 2.4 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 2.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.7 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 1.8 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 10.8 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 1.1 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.0 | 1.0 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 2.4 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 9.7 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.6 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 8.6 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.7 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.4 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 3.7 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 3.2 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 2.5 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 1.3 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 9.3 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 1.7 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.2 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 1.0 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 1.8 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.3 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.5 | 1.6 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.5 | 2.0 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.5 | 2.0 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.4 | 3.8 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.4 | 2.3 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.4 | 2.6 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.4 | 2.5 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.3 | 7.8 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.3 | 4.0 | GO:0015279 | store-operated calcium channel activity(GO:0015279) inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.3 | 1.0 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.2 | 4.5 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.2 | 4.6 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.2 | 1.6 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.2 | 1.9 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.2 | 0.7 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.2 | 1.5 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.2 | 0.7 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.2 | 1.3 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.2 | 1.1 | GO:0070191 | methionine-R-sulfoxide reductase activity(GO:0070191) |
| 0.1 | 1.0 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.1 | 10.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.7 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 2.2 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 0.8 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 0.8 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.1 | 0.6 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 1.2 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.1 | 0.6 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 0.4 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 1.2 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.1 | 1.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 1.7 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.6 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 1.6 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 3.3 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 1.0 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 0.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 1.7 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 1.3 | GO:0050253 | prenylcysteine methylesterase activity(GO:0010296) 1-oxa-2-oxocycloheptane lactonase activity(GO:0018731) sulfolactone hydrolase activity(GO:0018732) butyrolactone hydrolase activity(GO:0018734) endosulfan lactone lactonase activity(GO:0034892) L-ascorbate 6-phosphate lactonase activity(GO:0035460) Ser-tRNA(Thr) hydrolase activity(GO:0043905) Ala-tRNA(Pro) hydrolase activity(GO:0043906) Cys-tRNA(Pro) hydrolase activity(GO:0043907) Ser(Gly)-tRNA(Ala) hydrolase activity(GO:0043908) all-trans-retinyl-palmitate hydrolase, all-trans-retinol forming activity(GO:0047376) retinyl-palmitate esterase activity(GO:0050253) mannosyl-oligosaccharide 1,6-alpha-mannosidase activity(GO:0052767) mannosyl-oligosaccharide 1,3-alpha-mannosidase activity(GO:0052768) methyl indole-3-acetate esterase activity(GO:0080030) methyl salicylate esterase activity(GO:0080031) methyl jasmonate esterase activity(GO:0080032) |
| 0.1 | 0.4 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.4 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.2 | GO:0052833 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.2 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.3 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.5 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 1.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 1.2 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 2.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0001093 | TFIIB-class transcription factor binding(GO:0001093) |
| 0.0 | 2.4 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 1.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.8 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 1.6 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.6 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.6 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.8 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.3 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.1 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.1 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.6 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 1.8 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 1.3 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.4 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.2 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |