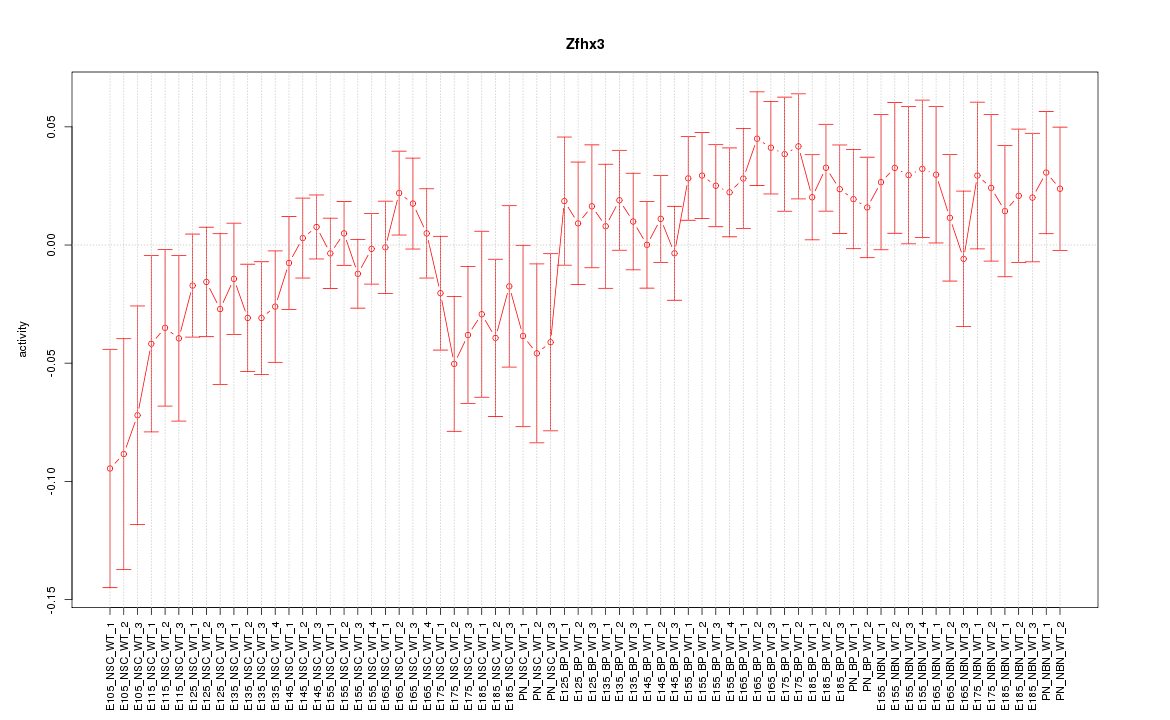
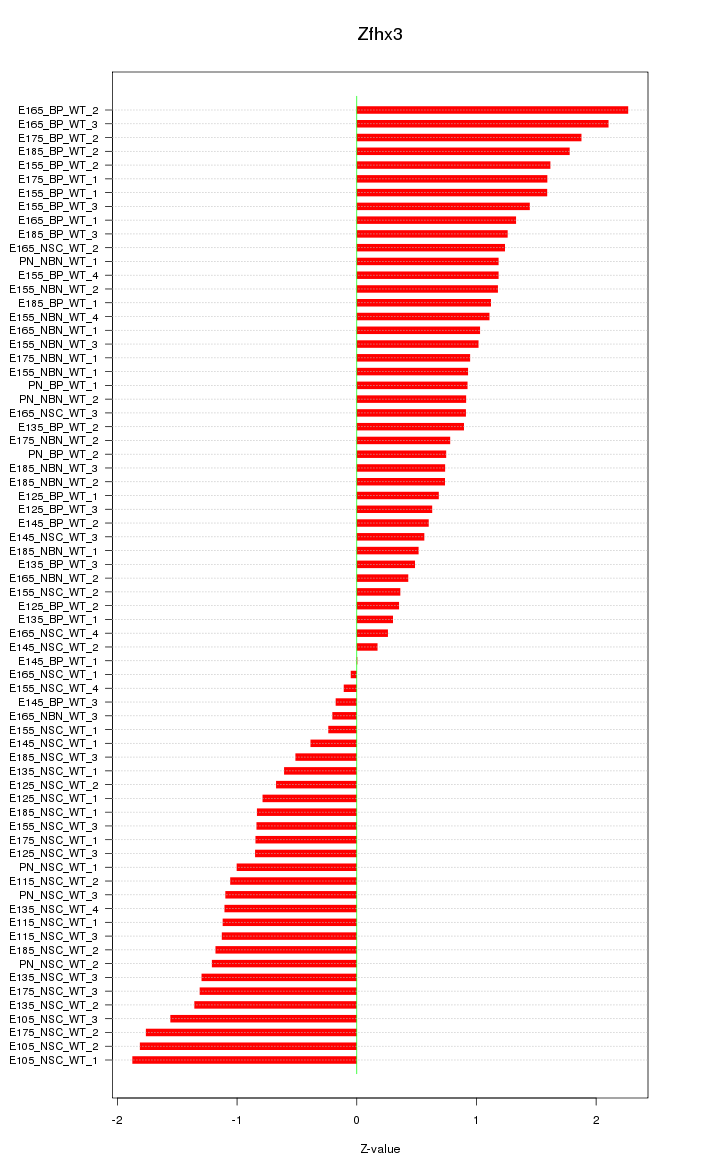
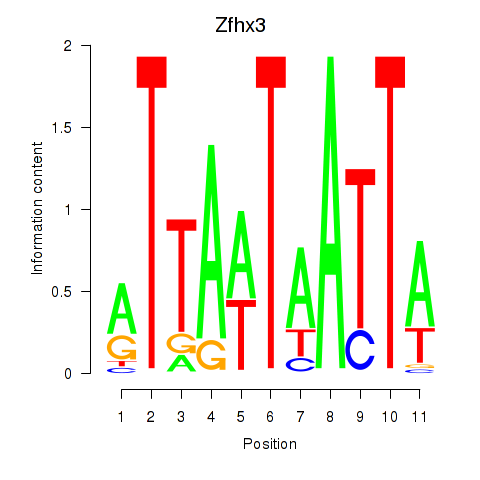
Motif ID: Zfhx3
Z-value: 1.083
Transcription factors associated with Zfhx3:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Zfhx3 | ENSMUSG00000038872.8 | Zfhx3 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfhx3 | mm10_v2_chr8_+_108714644_108714644 | -0.46 | 6.2e-05 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.5 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 2.1 | 6.2 | GO:0003345 | proepicardium cell migration involved in pericardium morphogenesis(GO:0003345) |
| 1.9 | 9.6 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 1.9 | 5.7 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 1.7 | 31.4 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 1.6 | 9.5 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 1.3 | 16.9 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.9 | 5.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.7 | 3.4 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.6 | 1.7 | GO:1904884 | telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.5 | 3.1 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.5 | 6.8 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.4 | 1.6 | GO:1900272 | negative regulation of long-term synaptic potentiation(GO:1900272) |
| 0.4 | 1.8 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.4 | 2.2 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.3 | 7.9 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.3 | 2.4 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.3 | 8.7 | GO:0095500 | acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.2 | 1.4 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.2 | 0.6 | GO:0042536 | negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.2 | 8.8 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.2 | 17.8 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.2 | 1.2 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.1 | 3.3 | GO:0048712 | negative regulation of astrocyte differentiation(GO:0048712) |
| 0.1 | 4.2 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.1 | 2.1 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 4.5 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.1 | 5.1 | GO:0036465 | synaptic vesicle recycling(GO:0036465) |
| 0.1 | 2.6 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 6.8 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.1 | 1.3 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 3.2 | GO:0009395 | phospholipid catabolic process(GO:0009395) |
| 0.1 | 3.1 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.1 | 0.6 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.1 | 1.5 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.1 | 2.8 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.1 | 7.6 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.0 | 1.9 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.0 | 0.4 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.0 | 0.5 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.2 | GO:0032929 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 1.9 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 3.9 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 3.2 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.5 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 3.6 | GO:0032496 | response to lipopolysaccharide(GO:0032496) |
| 0.0 | 0.8 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.2 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.9 | 5.7 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.4 | 3.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.4 | 2.2 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.4 | 3.4 | GO:0045275 | respiratory chain complex III(GO:0045275) |
| 0.3 | 1.2 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.2 | 7.5 | GO:0030673 | axolemma(GO:0030673) |
| 0.2 | 8.6 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.2 | 2.6 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.2 | 1.8 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 1.9 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 6.2 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 8.8 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 17.9 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 4.5 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 0.8 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 9.6 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.1 | 1.4 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 1.7 | GO:0001741 | XY body(GO:0001741) |
| 0.1 | 7.9 | GO:0042641 | actomyosin(GO:0042641) |
| 0.1 | 6.8 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 3.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 5.1 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 28.7 | GO:0005874 | microtubule(GO:0005874) |
| 0.1 | 3.9 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 1.6 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 13.8 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 9.6 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 2.8 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 9.0 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 12.5 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.6 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 8.7 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 1.3 | 31.4 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.9 | 5.7 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.9 | 3.6 | GO:0004954 | icosanoid receptor activity(GO:0004953) prostanoid receptor activity(GO:0004954) prostaglandin receptor activity(GO:0004955) |
| 0.8 | 7.5 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.8 | 17.9 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.6 | 6.8 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.5 | 3.1 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.4 | 3.4 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.4 | 7.6 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.4 | 5.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.4 | 2.2 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.3 | 1.6 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.3 | 6.2 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.3 | 1.7 | GO:0032405 | MutLalpha complex binding(GO:0032405) MutSalpha complex binding(GO:0032407) |
| 0.3 | 3.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.2 | 3.2 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.2 | 1.8 | GO:0004889 | acetylcholine-activated cation-selective channel activity(GO:0004889) |
| 0.1 | 2.8 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 1.2 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 3.9 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 2.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 16.9 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 9.6 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.1 | 7.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 9.5 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 6.5 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.1 | 4.1 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 8.3 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 8.8 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.6 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.4 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.9 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 1.9 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.1 | GO:0004984 | olfactory receptor activity(GO:0004984) |