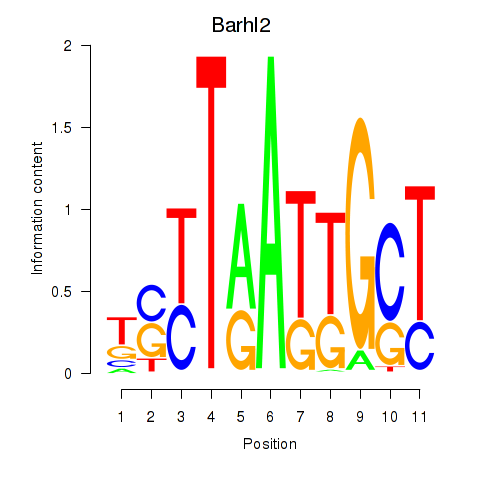
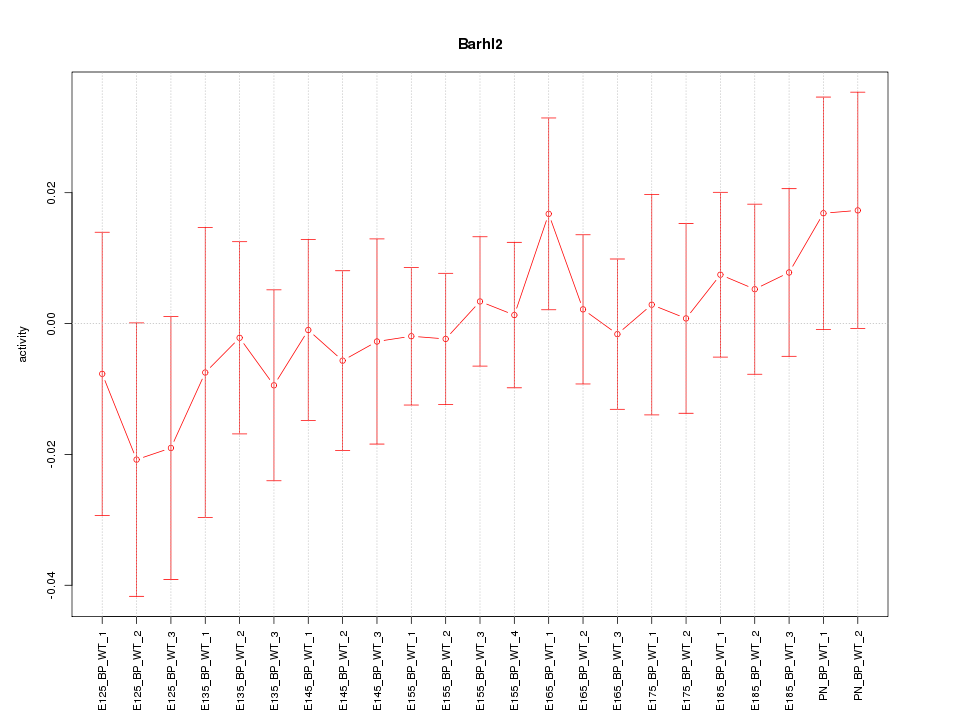
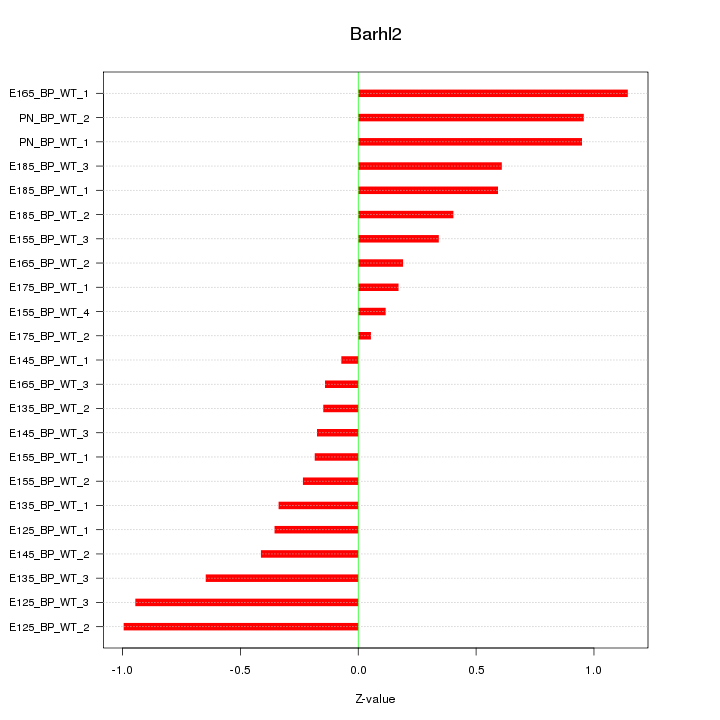
| 0.7 |
3.6 |
GO:0048133 |
germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.5 |
1.6 |
GO:0072223 |
metanephric glomerular mesangium development(GO:0072223) metanephric glomerular mesangial cell proliferation involved in metanephros development(GO:0072262) |
| 0.5 |
1.6 |
GO:0002447 |
eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) regulation of eosinophil activation(GO:1902566) |
| 0.3 |
1.0 |
GO:0007521 |
muscle cell fate determination(GO:0007521) mammary placode formation(GO:0060596) |
| 0.3 |
0.9 |
GO:0035880 |
embryonic nail plate morphogenesis(GO:0035880) |
| 0.3 |
0.9 |
GO:0008065 |
establishment of blood-nerve barrier(GO:0008065) regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.2 |
1.6 |
GO:0015808 |
L-alanine transport(GO:0015808) |
| 0.2 |
0.8 |
GO:1902990 |
mitotic telomere maintenance via semi-conservative replication(GO:1902990) negative regulation of t-circle formation(GO:1904430) |
| 0.2 |
1.9 |
GO:0034983 |
peptidyl-lysine deacetylation(GO:0034983) regulation of skeletal muscle fiber development(GO:0048742) |
| 0.2 |
3.1 |
GO:0016226 |
iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.2 |
1.1 |
GO:0015840 |
urea transport(GO:0015840) urea transmembrane transport(GO:0071918) |
| 0.2 |
0.8 |
GO:0070384 |
Harderian gland development(GO:0070384) |
| 0.1 |
1.1 |
GO:2000809 |
neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.1 |
0.5 |
GO:1903463 |
mitotic cell cycle phase(GO:0098763) regulation of mitotic cell cycle DNA replication(GO:1903463) |
| 0.1 |
0.6 |
GO:1903056 |
regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.1 |
0.4 |
GO:2000974 |
negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.1 |
2.0 |
GO:0035235 |
ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 |
0.4 |
GO:0046598 |
positive regulation of viral entry into host cell(GO:0046598) |
| 0.1 |
1.4 |
GO:0016339 |
calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 |
0.4 |
GO:0010025 |
wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.1 |
0.9 |
GO:0042761 |
very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.1 |
0.5 |
GO:0031119 |
tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 |
0.5 |
GO:0030579 |
ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 |
0.8 |
GO:0043252 |
sodium-independent organic anion transport(GO:0043252) |
| 0.1 |
0.2 |
GO:0016321 |
female meiosis chromosome segregation(GO:0016321) |
| 0.1 |
2.8 |
GO:0007528 |
neuromuscular junction development(GO:0007528) |
| 0.0 |
0.6 |
GO:0021891 |
olfactory bulb interneuron development(GO:0021891) |
| 0.0 |
0.8 |
GO:0097062 |
dendritic spine maintenance(GO:0097062) |
| 0.0 |
0.8 |
GO:0034453 |
microtubule anchoring(GO:0034453) |
| 0.0 |
2.6 |
GO:0050852 |
T cell receptor signaling pathway(GO:0050852) |
| 0.0 |
0.6 |
GO:0032287 |
peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 |
0.3 |
GO:1904153 |
negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 |
0.1 |
GO:0071034 |
CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 |
1.0 |
GO:0031365 |
N-terminal protein amino acid modification(GO:0031365) |
| 0.0 |
0.5 |
GO:0001833 |
inner cell mass cell proliferation(GO:0001833) |
| 0.0 |
0.1 |
GO:0031117 |
positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 |
0.4 |
GO:0030150 |
protein import into mitochondrial matrix(GO:0030150) |
| 0.0 |
0.2 |
GO:0060134 |
prepulse inhibition(GO:0060134) |
| 0.0 |
0.1 |
GO:0045144 |
meiotic sister chromatid segregation(GO:0045144) meiotic sister chromatid cohesion(GO:0051177) |
| 0.0 |
0.4 |
GO:0048305 |
immunoglobulin secretion(GO:0048305) |
| 0.0 |
0.3 |
GO:0031507 |
heterochromatin assembly(GO:0031507) |
| 0.0 |
0.7 |
GO:0002474 |
antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 |
0.3 |
GO:2000001 |
regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 |
0.1 |
GO:0016322 |
neuron remodeling(GO:0016322) |
| 0.0 |
0.5 |
GO:0016578 |
histone deubiquitination(GO:0016578) |
| 0.0 |
1.5 |
GO:0030168 |
platelet activation(GO:0030168) |
| 0.0 |
0.0 |
GO:1904742 |
protein poly-ADP-ribosylation(GO:0070212) regulation of telomeric DNA binding(GO:1904742) |
| 0.0 |
0.4 |
GO:0031114 |
negative regulation of microtubule depolymerization(GO:0007026) regulation of microtubule depolymerization(GO:0031114) |
| 0.0 |
0.3 |
GO:0032011 |
ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 |
0.3 |
GO:0000132 |
establishment of mitotic spindle orientation(GO:0000132) |