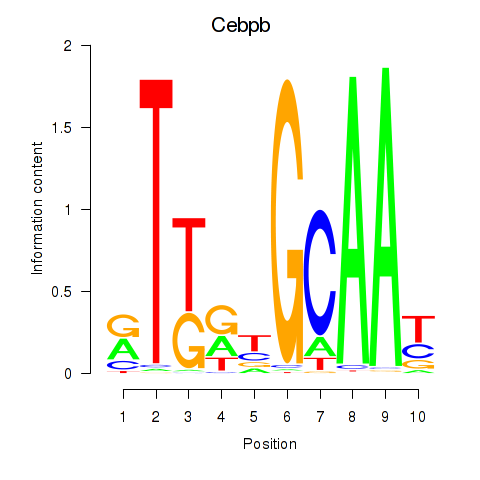
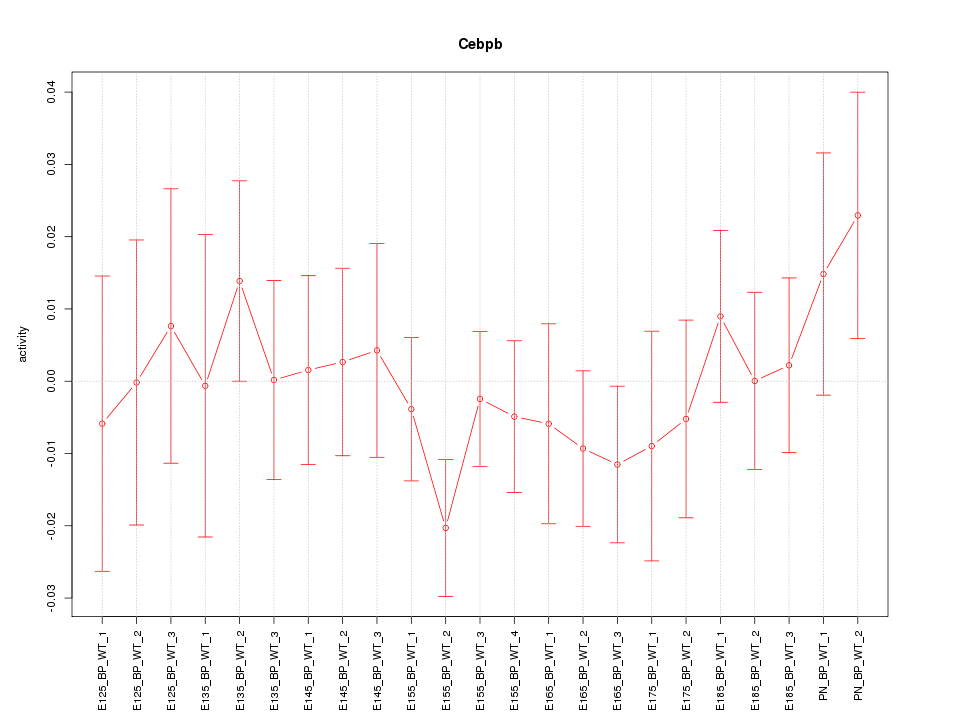
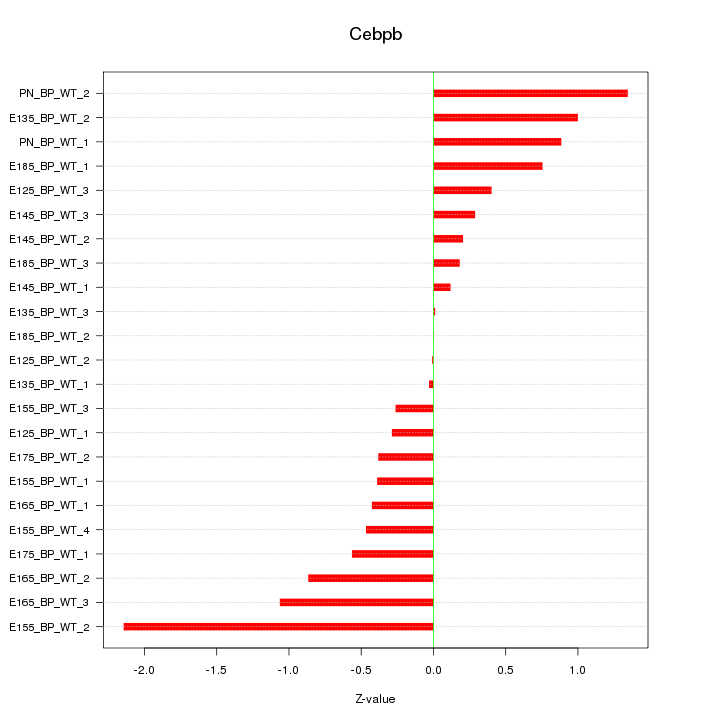
| 0.5 |
3.3 |
GO:1902847 |
macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of tau-protein kinase activity(GO:1902949) |
| 0.4 |
1.8 |
GO:2000255 |
negative regulation of male germ cell proliferation(GO:2000255) |
| 0.3 |
1.0 |
GO:1904706 |
response to cisplatin(GO:0072718) negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.3 |
0.8 |
GO:2000562 |
negative regulation of interferon-gamma secretion(GO:1902714) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.2 |
1.4 |
GO:0021797 |
forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.2 |
0.9 |
GO:0002580 |
regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002580) |
| 0.2 |
0.7 |
GO:0015920 |
lipopolysaccharide transport(GO:0015920) |
| 0.2 |
0.7 |
GO:0035790 |
platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.2 |
0.6 |
GO:0033364 |
mast cell secretory granule organization(GO:0033364) |
| 0.1 |
1.1 |
GO:0060770 |
negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.1 |
0.7 |
GO:0006564 |
L-serine biosynthetic process(GO:0006564) |
| 0.1 |
0.8 |
GO:1900747 |
negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 |
0.3 |
GO:0001580 |
detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 |
0.9 |
GO:0048251 |
elastic fiber assembly(GO:0048251) |
| 0.1 |
0.4 |
GO:0010808 |
positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.1 |
0.3 |
GO:1903984 |
positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.1 |
0.3 |
GO:0042167 |
heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.1 |
0.2 |
GO:0045659 |
regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.1 |
0.3 |
GO:0071726 |
response to diacyl bacterial lipopeptide(GO:0071724) cellular response to diacyl bacterial lipopeptide(GO:0071726) |
| 0.1 |
0.2 |
GO:0032430 |
positive regulation of phospholipase A2 activity(GO:0032430) |
| 0.1 |
0.4 |
GO:0089711 |
L-glutamate transmembrane transport(GO:0089711) |
| 0.1 |
0.4 |
GO:0001867 |
complement activation, lectin pathway(GO:0001867) |
| 0.1 |
0.5 |
GO:0097460 |
positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) ferrous iron import into cell(GO:0097460) |
| 0.1 |
0.3 |
GO:0097527 |
necroptotic signaling pathway(GO:0097527) |
| 0.1 |
0.5 |
GO:0031665 |
negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.1 |
0.2 |
GO:1903903 |
regulation of establishment of T cell polarity(GO:1903903) |
| 0.1 |
0.2 |
GO:0000715 |
nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.1 |
0.4 |
GO:0061002 |
negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 |
0.1 |
GO:0006421 |
asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 |
0.4 |
GO:0032020 |
ISG15-protein conjugation(GO:0032020) |
| 0.0 |
0.3 |
GO:1903352 |
ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.0 |
0.4 |
GO:0043615 |
astrocyte cell migration(GO:0043615) |
| 0.0 |
0.1 |
GO:0060024 |
musculoskeletal movement, spinal reflex action(GO:0050883) rhythmic synaptic transmission(GO:0060024) |
| 0.0 |
0.2 |
GO:0018364 |
peptidyl-glutamine methylation(GO:0018364) |
| 0.0 |
0.6 |
GO:0034501 |
protein localization to kinetochore(GO:0034501) |
| 0.0 |
0.1 |
GO:0018343 |
protein farnesylation(GO:0018343) |
| 0.0 |
0.2 |
GO:0046689 |
response to mercury ion(GO:0046689) |
| 0.0 |
0.1 |
GO:2000620 |
positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.0 |
0.1 |
GO:0060217 |
hemangioblast cell differentiation(GO:0060217) |
| 0.0 |
0.4 |
GO:0046688 |
response to copper ion(GO:0046688) |
| 0.0 |
0.1 |
GO:0021759 |
globus pallidus development(GO:0021759) |
| 0.0 |
0.3 |
GO:0045187 |
regulation of circadian sleep/wake cycle(GO:0042749) regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.0 |
0.3 |
GO:0044539 |
long-chain fatty acid import(GO:0044539) |
| 0.0 |
0.1 |
GO:0046122 |
purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.0 |
0.6 |
GO:0046185 |
aldehyde catabolic process(GO:0046185) |
| 0.0 |
2.2 |
GO:0003073 |
regulation of systemic arterial blood pressure(GO:0003073) |
| 0.0 |
0.2 |
GO:0046085 |
adenosine metabolic process(GO:0046085) |
| 0.0 |
0.1 |
GO:0072675 |
osteoclast fusion(GO:0072675) |
| 0.0 |
0.5 |
GO:0060391 |
positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 |
0.1 |
GO:0060390 |
regulation of SMAD protein import into nucleus(GO:0060390) |
| 0.0 |
0.2 |
GO:0002087 |
regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 |
0.1 |
GO:0002934 |
desmosome organization(GO:0002934) |
| 0.0 |
0.2 |
GO:0045109 |
intermediate filament organization(GO:0045109) |
| 0.0 |
0.4 |
GO:0043552 |
positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 |
0.6 |
GO:0050819 |
negative regulation of coagulation(GO:0050819) |
| 0.0 |
0.1 |
GO:1990086 |
lens fiber cell apoptotic process(GO:1990086) |
| 0.0 |
0.1 |
GO:0046477 |
glycosylceramide catabolic process(GO:0046477) |
| 0.0 |
0.9 |
GO:0045744 |
negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 |
0.1 |
GO:0045876 |
positive regulation of sister chromatid cohesion(GO:0045876) positive regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902177) |
| 0.0 |
0.4 |
GO:0032981 |
NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 |
0.1 |
GO:0072592 |
oxygen metabolic process(GO:0072592) |
| 0.0 |
0.2 |
GO:0038166 |
angiotensin-activated signaling pathway(GO:0038166) |
| 0.0 |
0.3 |
GO:0090129 |
positive regulation of synapse maturation(GO:0090129) |
| 0.0 |
0.1 |
GO:0016259 |
selenocysteine metabolic process(GO:0016259) |
| 0.0 |
0.2 |
GO:0009048 |
dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 |
0.4 |
GO:0040034 |
regulation of development, heterochronic(GO:0040034) |
| 0.0 |
0.3 |
GO:0002495 |
antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.0 |
0.1 |
GO:0015808 |
L-alanine transport(GO:0015808) |
| 0.0 |
1.3 |
GO:0031638 |
zymogen activation(GO:0031638) |
| 0.0 |
0.1 |
GO:0046485 |
ether lipid metabolic process(GO:0046485) |
| 0.0 |
0.3 |
GO:0042249 |
establishment of planar polarity of embryonic epithelium(GO:0042249) |
| 0.0 |
0.1 |
GO:0006049 |
UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 |
0.0 |
GO:0090202 |
transcriptional activation by promoter-enhancer looping(GO:0071733) regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.0 |
0.3 |
GO:0043171 |
peptide catabolic process(GO:0043171) |
| 0.0 |
0.2 |
GO:0034389 |
lipid particle organization(GO:0034389) |
| 0.0 |
0.4 |
GO:0036075 |
endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |