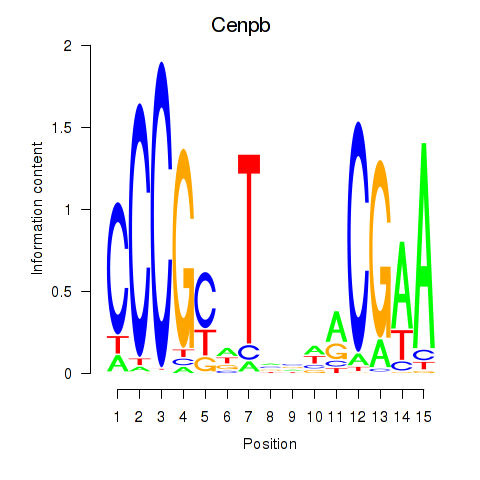
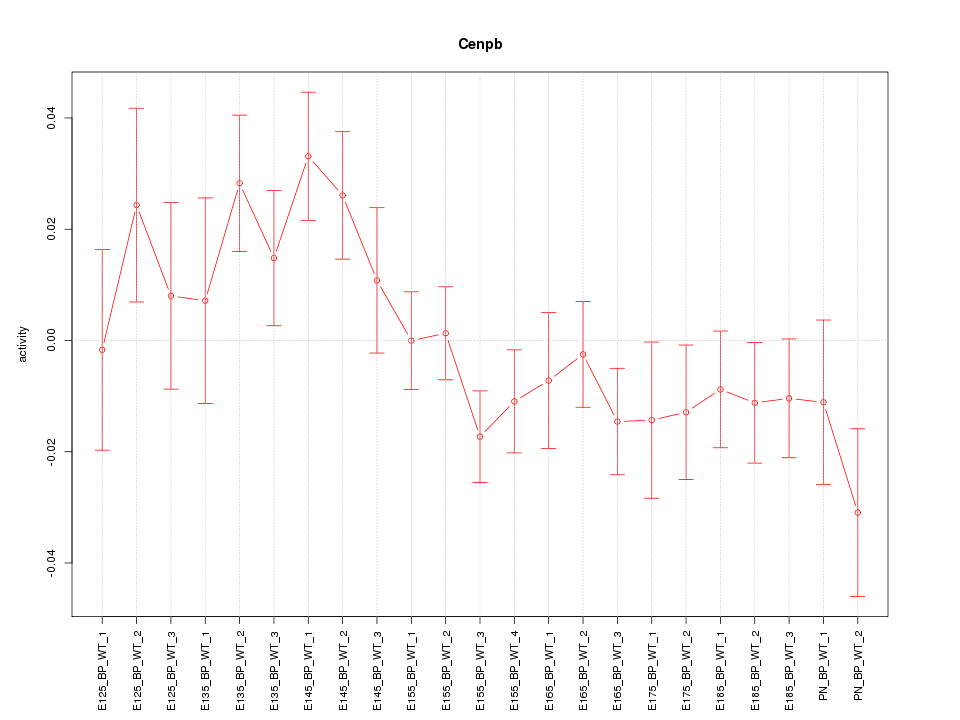
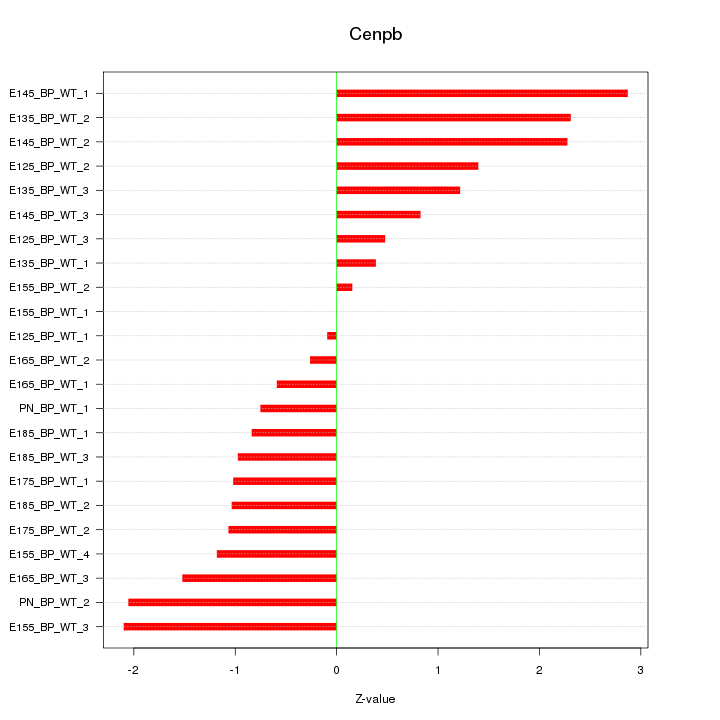
| 1.4 |
4.1 |
GO:0060598 |
dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 0.8 |
3.2 |
GO:2000546 |
positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.6 |
2.9 |
GO:0035262 |
gonad morphogenesis(GO:0035262) |
| 0.6 |
0.6 |
GO:2000830 |
vacuolar phosphate transport(GO:0007037) vitamin D3 metabolic process(GO:0070640) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 0.5 |
1.4 |
GO:2001206 |
positive regulation of osteoclast development(GO:2001206) |
| 0.4 |
5.6 |
GO:0042711 |
maternal behavior(GO:0042711) |
| 0.4 |
1.2 |
GO:0032474 |
otolith morphogenesis(GO:0032474) |
| 0.4 |
1.5 |
GO:0044330 |
canonical Wnt signaling pathway involved in positive regulation of wound healing(GO:0044330) lactic acid secretion(GO:0046722) regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.3 |
4.9 |
GO:0060052 |
neurofilament cytoskeleton organization(GO:0060052) |
| 0.3 |
1.3 |
GO:0010940 |
positive regulation of necrotic cell death(GO:0010940) |
| 0.3 |
1.2 |
GO:1903237 |
negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.3 |
1.1 |
GO:0002035 |
brain renin-angiotensin system(GO:0002035) positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.3 |
0.8 |
GO:2000832 |
protein-chromophore linkage(GO:0018298) negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.3 |
3.1 |
GO:0051601 |
exocyst localization(GO:0051601) |
| 0.3 |
0.5 |
GO:0042726 |
flavin-containing compound metabolic process(GO:0042726) |
| 0.2 |
0.7 |
GO:0002669 |
positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.2 |
1.5 |
GO:1900016 |
negative regulation of cytokine production involved in inflammatory response(GO:1900016) |
| 0.2 |
1.7 |
GO:1903715 |
regulation of aerobic respiration(GO:1903715) |
| 0.2 |
0.7 |
GO:2000620 |
positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.2 |
0.7 |
GO:0001978 |
regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) |
| 0.2 |
0.4 |
GO:0072076 |
nephrogenic mesenchyme development(GO:0072076) kidney mesenchyme morphogenesis(GO:0072131) metanephric mesenchyme morphogenesis(GO:0072133) |
| 0.2 |
0.9 |
GO:0033499 |
galactose catabolic process via UDP-galactose(GO:0033499) glycolytic process from galactose(GO:0061623) |
| 0.2 |
1.5 |
GO:1904667 |
negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.2 |
1.1 |
GO:0034436 |
glycoprotein transport(GO:0034436) |
| 0.2 |
0.6 |
GO:0036388 |
pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.2 |
0.6 |
GO:0033615 |
mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.2 |
0.6 |
GO:0031038 |
meiotic chromosome movement towards spindle pole(GO:0016344) myosin II filament organization(GO:0031038) regulation of myosin II filament organization(GO:0043519) |
| 0.2 |
0.8 |
GO:0060266 |
negative regulation of respiratory burst involved in inflammatory response(GO:0060266) negative regulation of respiratory burst(GO:0060268) |
| 0.2 |
0.6 |
GO:1903334 |
positive regulation of protein folding(GO:1903334) |
| 0.2 |
2.0 |
GO:0033089 |
positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.2 |
0.9 |
GO:0070459 |
prolactin secretion(GO:0070459) |
| 0.2 |
0.7 |
GO:1990414 |
replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.2 |
1.5 |
GO:1903298 |
regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903297) negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903298) |
| 0.2 |
0.5 |
GO:0000720 |
pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.2 |
0.6 |
GO:0060741 |
prostate gland stromal morphogenesis(GO:0060741) |
| 0.2 |
0.5 |
GO:0010040 |
response to iron(II) ion(GO:0010040) |
| 0.1 |
1.3 |
GO:0048386 |
positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 |
0.8 |
GO:0071712 |
ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 |
3.9 |
GO:0006879 |
cellular iron ion homeostasis(GO:0006879) |
| 0.1 |
0.6 |
GO:0070327 |
thyroid hormone transport(GO:0070327) |
| 0.1 |
0.9 |
GO:0061002 |
negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.1 |
0.7 |
GO:0072610 |
interleukin-12 secretion(GO:0072610) regulation of interleukin-12 secretion(GO:2001182) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.1 |
0.5 |
GO:0046709 |
IDP metabolic process(GO:0046707) IDP catabolic process(GO:0046709) |
| 0.1 |
1.4 |
GO:0071257 |
cellular response to electrical stimulus(GO:0071257) |
| 0.1 |
0.6 |
GO:0006452 |
translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 |
0.8 |
GO:0036289 |
peptidyl-serine autophosphorylation(GO:0036289) |
| 0.1 |
0.7 |
GO:0044027 |
hypermethylation of CpG island(GO:0044027) |
| 0.1 |
0.4 |
GO:0007603 |
phototransduction, visible light(GO:0007603) |
| 0.1 |
1.0 |
GO:0031274 |
positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 |
1.4 |
GO:0032515 |
negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 |
1.5 |
GO:0045653 |
negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.1 |
0.6 |
GO:2000786 |
positive regulation of autophagosome assembly(GO:2000786) |
| 0.1 |
2.1 |
GO:0097320 |
membrane tubulation(GO:0097320) |
| 0.1 |
0.4 |
GO:0006680 |
glucosylceramide catabolic process(GO:0006680) |
| 0.1 |
0.4 |
GO:0006975 |
DNA damage induced protein phosphorylation(GO:0006975) regulation of mitotic centrosome separation(GO:0046602) |
| 0.1 |
0.2 |
GO:0002572 |
pro-T cell differentiation(GO:0002572) ureter maturation(GO:0035799) cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) regulation of DNA demethylation(GO:1901535) negative regulation of DNA demethylation(GO:1901536) |
| 0.1 |
1.2 |
GO:0016226 |
iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 |
0.3 |
GO:0015744 |
succinate transport(GO:0015744) |
| 0.1 |
0.7 |
GO:2000628 |
regulation of miRNA metabolic process(GO:2000628) |
| 0.1 |
0.4 |
GO:1901524 |
regulation of macromitophagy(GO:1901524) negative regulation of macromitophagy(GO:1901525) negative regulation of mitophagy(GO:1903147) |
| 0.1 |
0.5 |
GO:0070445 |
oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.1 |
1.1 |
GO:1902187 |
negative regulation of viral release from host cell(GO:1902187) |
| 0.1 |
0.4 |
GO:0000055 |
ribosomal large subunit export from nucleus(GO:0000055) |
| 0.1 |
0.2 |
GO:0006407 |
rRNA export from nucleus(GO:0006407) |
| 0.1 |
0.7 |
GO:0006268 |
DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 |
1.3 |
GO:0060716 |
labyrinthine layer blood vessel development(GO:0060716) |
| 0.1 |
0.2 |
GO:0034310 |
primary alcohol catabolic process(GO:0034310) |
| 0.1 |
0.7 |
GO:0006654 |
phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.1 |
0.6 |
GO:0010216 |
maintenance of DNA methylation(GO:0010216) |
| 0.1 |
0.6 |
GO:0021942 |
radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.1 |
0.4 |
GO:0014049 |
positive regulation of glutamate secretion(GO:0014049) |
| 0.0 |
1.1 |
GO:0007205 |
protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 |
0.5 |
GO:0006995 |
cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 |
0.2 |
GO:0031053 |
primary miRNA processing(GO:0031053) |
| 0.0 |
0.8 |
GO:0006999 |
nuclear pore organization(GO:0006999) |
| 0.0 |
1.4 |
GO:0030513 |
positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 |
0.1 |
GO:0071316 |
cellular response to nicotine(GO:0071316) |
| 0.0 |
1.1 |
GO:0018345 |
protein palmitoylation(GO:0018345) |
| 0.0 |
0.2 |
GO:0051697 |
protein delipidation(GO:0051697) |
| 0.0 |
0.2 |
GO:0046600 |
negative regulation of centriole replication(GO:0046600) |
| 0.0 |
0.7 |
GO:0043248 |
proteasome assembly(GO:0043248) |
| 0.0 |
0.7 |
GO:0046069 |
cGMP catabolic process(GO:0046069) |
| 0.0 |
0.5 |
GO:0030150 |
protein import into mitochondrial matrix(GO:0030150) |
| 0.0 |
0.1 |
GO:0042796 |
snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 |
0.5 |
GO:0015937 |
coenzyme A biosynthetic process(GO:0015937) |
| 0.0 |
0.3 |
GO:0010815 |
bradykinin catabolic process(GO:0010815) |
| 0.0 |
0.9 |
GO:0008299 |
isoprenoid biosynthetic process(GO:0008299) |
| 0.0 |
0.1 |
GO:0038128 |
ERBB2 signaling pathway(GO:0038128) |
| 0.0 |
1.5 |
GO:0005978 |
glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 |
0.1 |
GO:0034143 |
regulation of toll-like receptor 4 signaling pathway(GO:0034143) |
| 0.0 |
1.2 |
GO:2001222 |
regulation of neuron migration(GO:2001222) |
| 0.0 |
0.3 |
GO:0034244 |
negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 |
0.4 |
GO:0016973 |
poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 |
0.4 |
GO:0043584 |
nose development(GO:0043584) |
| 0.0 |
0.2 |
GO:0048742 |
regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 |
1.4 |
GO:0030512 |
negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 |
0.2 |
GO:0017196 |
N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 |
0.1 |
GO:0060242 |
contact inhibition(GO:0060242) |
| 0.0 |
1.2 |
GO:0070303 |
negative regulation of stress-activated MAPK cascade(GO:0032873) negative regulation of stress-activated protein kinase signaling cascade(GO:0070303) |
| 0.0 |
0.3 |
GO:1900119 |
positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 |
0.1 |
GO:0006106 |
fumarate metabolic process(GO:0006106) |
| 0.0 |
0.9 |
GO:0007193 |
adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 |
0.4 |
GO:0051123 |
RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 |
1.1 |
GO:0032543 |
mitochondrial translation(GO:0032543) |
| 0.0 |
0.8 |
GO:1902653 |
cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 |
0.7 |
GO:0001947 |
heart looping(GO:0001947) |
| 0.0 |
0.1 |
GO:0007135 |
meiosis II(GO:0007135) meiotic sister chromatid segregation(GO:0045144) meiotic sister chromatid cohesion(GO:0051177) |
| 0.0 |
1.4 |
GO:0010950 |
positive regulation of endopeptidase activity(GO:0010950) |
| 0.0 |
0.1 |
GO:0033540 |
fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 |
0.0 |
GO:0035552 |
oxidative single-stranded DNA demethylation(GO:0035552) |
| 0.0 |
0.8 |
GO:0009060 |
aerobic respiration(GO:0009060) |
| 0.0 |
0.4 |
GO:0010922 |
positive regulation of phosphatase activity(GO:0010922) |
| 0.0 |
0.1 |
GO:1901387 |
positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.0 |
0.3 |
GO:1902259 |
regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.0 |
0.2 |
GO:0046716 |
muscle cell cellular homeostasis(GO:0046716) |
| 0.0 |
0.2 |
GO:0061157 |
mRNA destabilization(GO:0061157) |