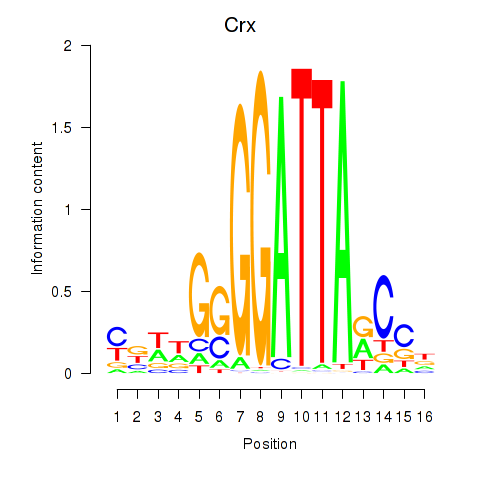
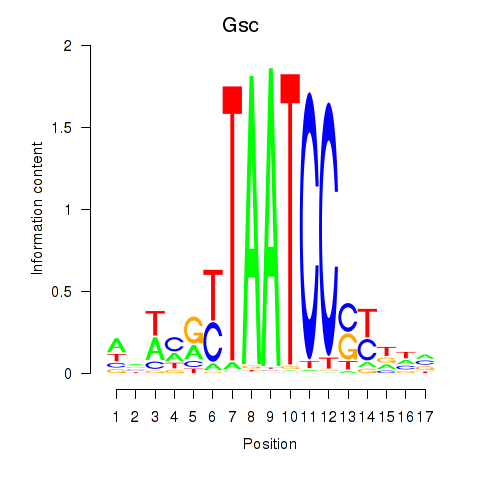
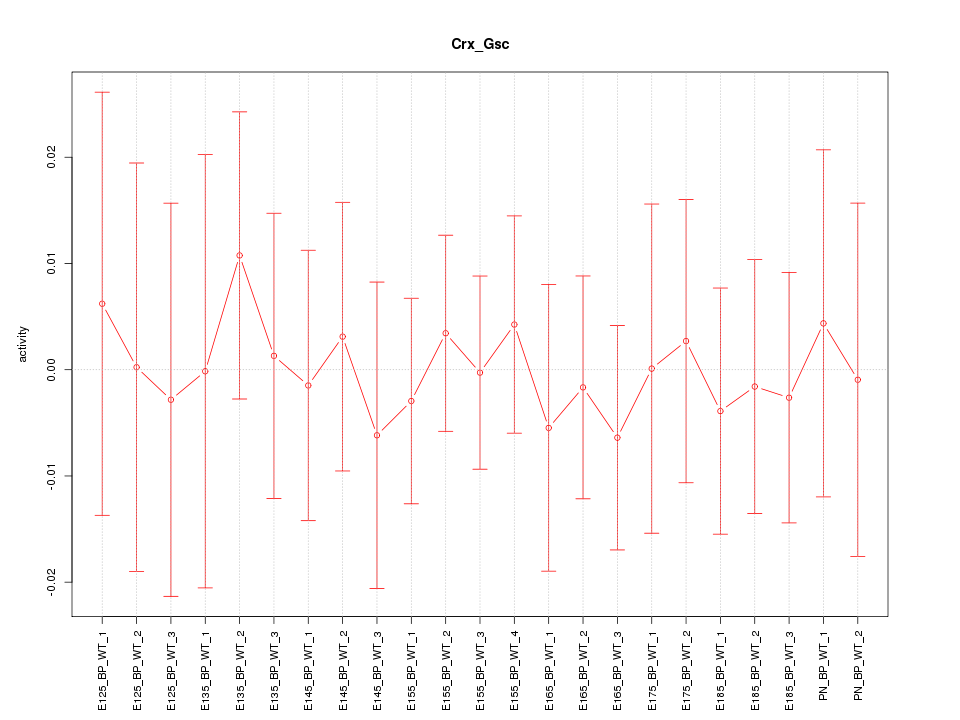
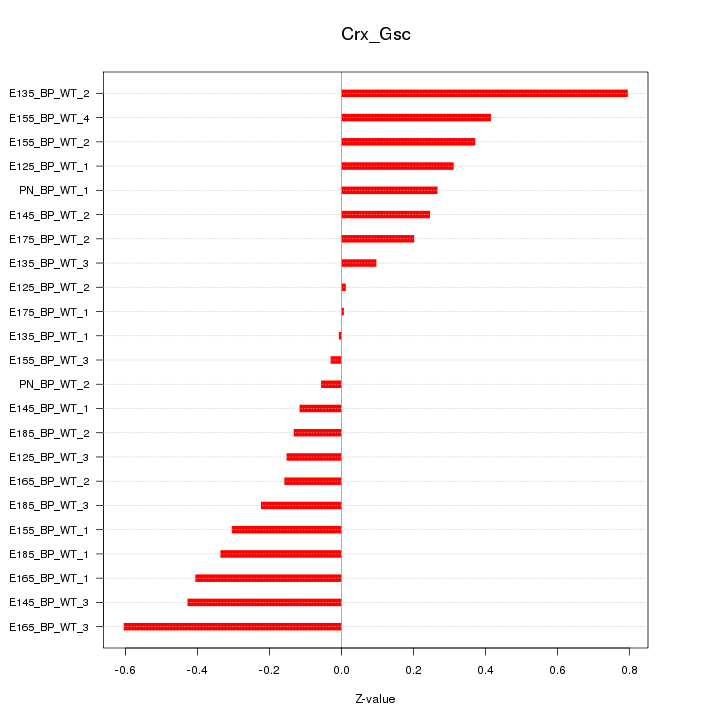
| 0.1 |
0.7 |
GO:0015671 |
oxygen transport(GO:0015671) |
| 0.1 |
0.4 |
GO:2000705 |
regulation of dense core granule biogenesis(GO:2000705) |
| 0.1 |
0.3 |
GO:0035262 |
gonad morphogenesis(GO:0035262) |
| 0.1 |
0.9 |
GO:0001731 |
formation of translation preinitiation complex(GO:0001731) |
| 0.1 |
0.1 |
GO:0072076 |
nephrogenic mesenchyme development(GO:0072076) |
| 0.0 |
0.1 |
GO:0050748 |
negative regulation of lipoprotein metabolic process(GO:0050748) |
| 0.0 |
0.1 |
GO:0060598 |
dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 0.0 |
0.1 |
GO:0032240 |
RNA import into nucleus(GO:0006404) mRNA export from nucleus in response to heat stress(GO:0031990) negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.0 |
0.2 |
GO:0090273 |
regulation of somatostatin secretion(GO:0090273) |
| 0.0 |
0.3 |
GO:1903608 |
protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 |
0.1 |
GO:0002669 |
positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 |
0.2 |
GO:0008343 |
adult feeding behavior(GO:0008343) |
| 0.0 |
0.1 |
GO:0010958 |
regulation of amino acid import(GO:0010958) regulation of L-arginine import(GO:0010963) |
| 0.0 |
0.1 |
GO:0002295 |
T-helper cell lineage commitment(GO:0002295) interleukin-4-mediated signaling pathway(GO:0035771) CD4-positive, alpha-beta T cell lineage commitment(GO:0043373) isotype switching to IgE isotypes(GO:0048289) regulation of isotype switching to IgE isotypes(GO:0048293) |
| 0.0 |
0.2 |
GO:0090209 |
negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.0 |
0.5 |
GO:0061000 |
negative regulation of dendritic spine development(GO:0061000) |
| 0.0 |
0.1 |
GO:0070315 |
G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.0 |
0.1 |
GO:0071442 |
positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 |
0.2 |
GO:2000348 |
regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 |
0.1 |
GO:0097461 |
ferric iron import(GO:0033216) ferric iron import into cell(GO:0097461) |
| 0.0 |
0.1 |
GO:1902998 |
macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of tau-protein kinase activity(GO:1902949) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.0 |
0.1 |
GO:0089711 |
L-glutamate transmembrane transport(GO:0089711) |
| 0.0 |
0.1 |
GO:0010920 |
negative regulation of inositol phosphate biosynthetic process(GO:0010920) |
| 0.0 |
0.1 |
GO:1990035 |
calcium ion import into cell(GO:1990035) |
| 0.0 |
0.1 |
GO:0019919 |
peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 |
0.1 |
GO:0006384 |
transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 |
0.1 |
GO:0048143 |
astrocyte activation(GO:0048143) |
| 0.0 |
0.0 |
GO:0045819 |
positive regulation of glycogen catabolic process(GO:0045819) |
| 0.0 |
0.1 |
GO:0071787 |
endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 |
0.1 |
GO:0002710 |
negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 |
0.1 |
GO:0003383 |
apical constriction(GO:0003383) |
| 0.0 |
0.1 |
GO:0006452 |
translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 |
0.1 |
GO:0048386 |
positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 |
0.1 |
GO:0030322 |
stabilization of membrane potential(GO:0030322) |
| 0.0 |
0.1 |
GO:0044458 |
motile cilium assembly(GO:0044458) |
| 0.0 |
0.1 |
GO:1900383 |
regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 |
0.0 |
GO:0038203 |
TORC2 signaling(GO:0038203) |
| 0.0 |
0.2 |
GO:0048268 |
clathrin coat assembly(GO:0048268) |