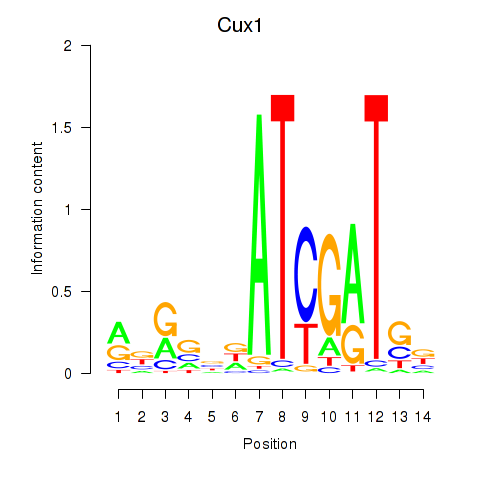
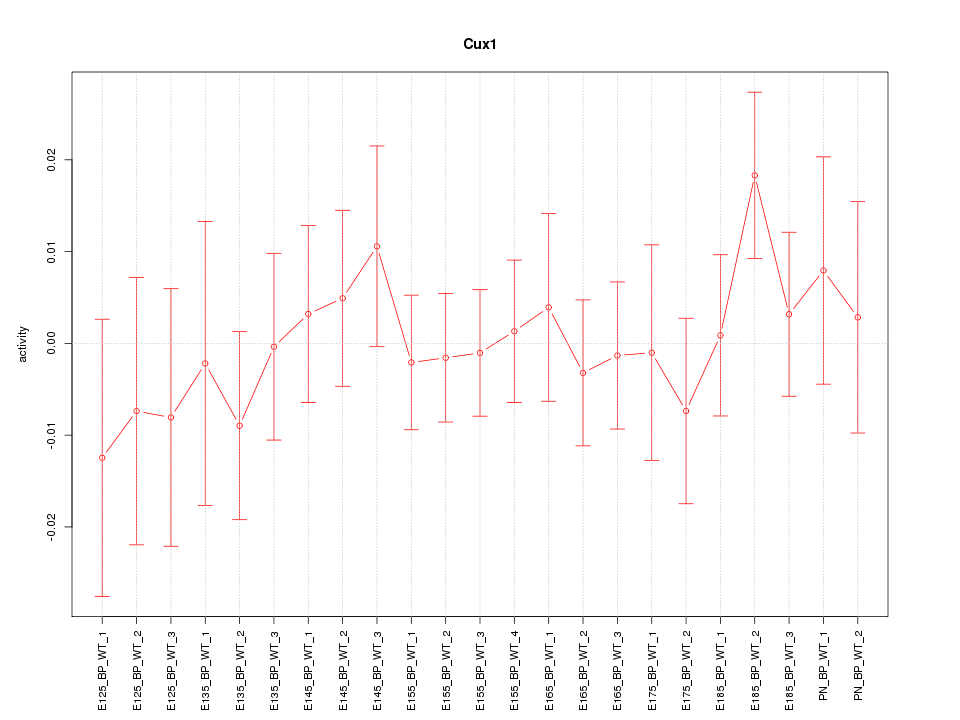
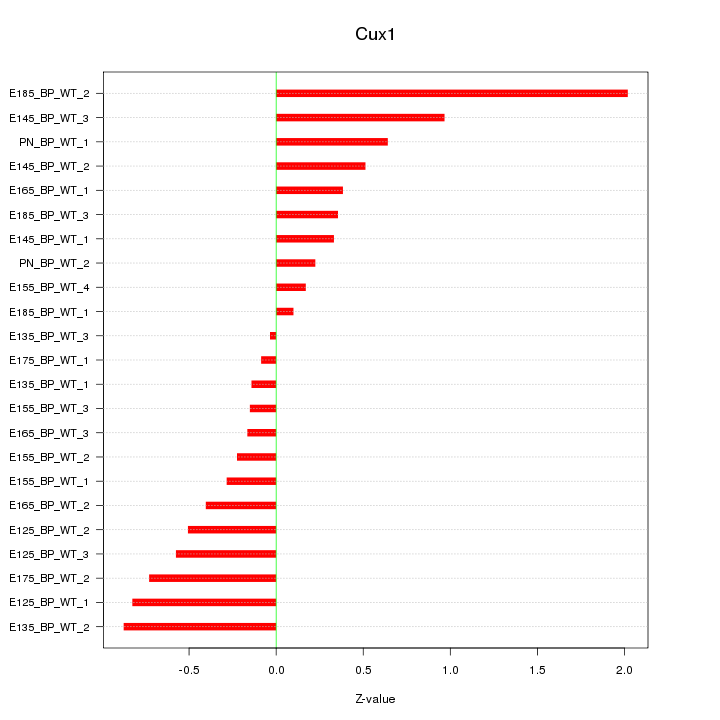
| 1.0 |
3.1 |
GO:0021893 |
cerebral cortex GABAergic interneuron fate commitment(GO:0021893) |
| 0.5 |
2.2 |
GO:1901204 |
positive regulation of guanylate cyclase activity(GO:0031284) regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) |
| 0.5 |
1.6 |
GO:2001139 |
negative regulation of postsynaptic membrane organization(GO:1901627) negative regulation of dendritic spine maintenance(GO:1902951) negative regulation of phospholipid efflux(GO:1902999) regulation of lipid transport across blood brain barrier(GO:1903000) negative regulation of lipid transport across blood brain barrier(GO:1903001) positive regulation of lipid transport across blood brain barrier(GO:1903002) negative regulation of phospholipid transport(GO:2001139) |
| 0.4 |
1.2 |
GO:1900041 |
intestinal D-glucose absorption(GO:0001951) negative regulation of interleukin-2 secretion(GO:1900041) terminal web assembly(GO:1902896) |
| 0.3 |
3.5 |
GO:0061302 |
smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.3 |
1.0 |
GO:0032289 |
central nervous system myelin formation(GO:0032289) |
| 0.3 |
0.5 |
GO:0034372 |
very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.2 |
0.5 |
GO:2001180 |
negative regulation of interleukin-18 production(GO:0032701) negative regulation of interleukin-10 secretion(GO:2001180) |
| 0.1 |
0.4 |
GO:0035622 |
intrahepatic bile duct development(GO:0035622) |
| 0.1 |
0.6 |
GO:2000525 |
regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.1 |
0.4 |
GO:0090403 |
oxidative stress-induced premature senescence(GO:0090403) |
| 0.1 |
0.6 |
GO:0006538 |
glutamate catabolic process(GO:0006538) |
| 0.1 |
0.4 |
GO:0046900 |
tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.1 |
1.6 |
GO:0034497 |
protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 |
0.6 |
GO:0032497 |
detection of lipopolysaccharide(GO:0032497) |
| 0.1 |
1.0 |
GO:0071569 |
protein ufmylation(GO:0071569) |
| 0.1 |
0.3 |
GO:0002447 |
eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) |
| 0.1 |
0.3 |
GO:0009182 |
purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) dGDP metabolic process(GO:0046066) GDP metabolic process(GO:0046710) |
| 0.1 |
0.4 |
GO:0045054 |
constitutive secretory pathway(GO:0045054) |
| 0.1 |
0.3 |
GO:0097155 |
fasciculation of sensory neuron axon(GO:0097155) |
| 0.1 |
0.1 |
GO:0042420 |
dopamine catabolic process(GO:0042420) |
| 0.1 |
0.3 |
GO:1900369 |
negative regulation of RNA interference(GO:1900369) |
| 0.1 |
0.3 |
GO:0046077 |
dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) deoxyribonucleoside diphosphate biosynthetic process(GO:0009189) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 0.1 |
0.4 |
GO:0046208 |
spermine catabolic process(GO:0046208) |
| 0.1 |
0.2 |
GO:0001869 |
regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.1 |
0.4 |
GO:0016127 |
cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 |
0.4 |
GO:0070295 |
renal water absorption(GO:0070295) |
| 0.1 |
0.2 |
GO:0006556 |
S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 |
0.6 |
GO:0045743 |
positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.1 |
0.6 |
GO:0035092 |
sperm chromatin condensation(GO:0035092) |
| 0.1 |
0.2 |
GO:2000983 |
regulation of ATP citrate synthase activity(GO:2000983) negative regulation of ATP citrate synthase activity(GO:2000984) |
| 0.1 |
0.2 |
GO:0002940 |
tRNA N2-guanine methylation(GO:0002940) |
| 0.1 |
0.1 |
GO:0070885 |
negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.1 |
0.6 |
GO:0071420 |
cellular response to histamine(GO:0071420) |
| 0.1 |
0.8 |
GO:0035269 |
protein O-linked mannosylation(GO:0035269) |
| 0.1 |
0.2 |
GO:1903279 |
regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.1 |
0.3 |
GO:0051541 |
elastin metabolic process(GO:0051541) |
| 0.1 |
0.2 |
GO:0006435 |
threonyl-tRNA aminoacylation(GO:0006435) |
| 0.1 |
0.1 |
GO:0035434 |
copper ion transmembrane transport(GO:0035434) |
| 0.1 |
0.3 |
GO:1902255 |
positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 0.1 |
0.2 |
GO:0015803 |
branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.1 |
0.3 |
GO:0060576 |
intestinal epithelial cell development(GO:0060576) |
| 0.1 |
0.3 |
GO:0032237 |
activation of store-operated calcium channel activity(GO:0032237) |
| 0.0 |
0.2 |
GO:0001920 |
negative regulation of receptor recycling(GO:0001920) |
| 0.0 |
0.5 |
GO:0015721 |
bile acid and bile salt transport(GO:0015721) |
| 0.0 |
0.3 |
GO:2000124 |
regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 |
0.2 |
GO:0019236 |
response to pheromone(GO:0019236) |
| 0.0 |
0.2 |
GO:0021508 |
floor plate formation(GO:0021508) floor plate morphogenesis(GO:0033505) |
| 0.0 |
1.0 |
GO:0007614 |
short-term memory(GO:0007614) |
| 0.0 |
0.7 |
GO:0050901 |
leukocyte tethering or rolling(GO:0050901) |
| 0.0 |
0.5 |
GO:0015816 |
glycine transport(GO:0015816) |
| 0.0 |
0.1 |
GO:0000389 |
generation of catalytic spliceosome for second transesterification step(GO:0000350) mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 |
0.1 |
GO:0009174 |
UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.0 |
0.7 |
GO:0031340 |
positive regulation of vesicle fusion(GO:0031340) |
| 0.0 |
0.2 |
GO:0006384 |
transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 |
0.1 |
GO:0048294 |
negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 |
0.1 |
GO:0006030 |
chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 |
0.8 |
GO:0019835 |
cytolysis(GO:0019835) |
| 0.0 |
0.1 |
GO:0035880 |
embryonic nail plate morphogenesis(GO:0035880) |
| 0.0 |
0.8 |
GO:0007214 |
gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 |
0.3 |
GO:0006621 |
protein retention in ER lumen(GO:0006621) |
| 0.0 |
0.1 |
GO:0006436 |
tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 |
0.6 |
GO:0060707 |
trophoblast giant cell differentiation(GO:0060707) |
| 0.0 |
0.3 |
GO:0070813 |
hydrogen sulfide metabolic process(GO:0070813) |
| 0.0 |
0.2 |
GO:1904721 |
regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) negative regulation of immunoglobulin secretion(GO:0051025) negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.0 |
0.2 |
GO:0006072 |
glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 |
0.2 |
GO:0006689 |
ganglioside catabolic process(GO:0006689) |
| 0.0 |
0.1 |
GO:1900272 |
regulation of chronic inflammatory response(GO:0002676) negative regulation of chronic inflammatory response(GO:0002677) negative regulation of long-term synaptic potentiation(GO:1900272) |
| 0.0 |
0.2 |
GO:0042135 |
neurotransmitter catabolic process(GO:0042135) |
| 0.0 |
0.3 |
GO:0018095 |
protein polyglutamylation(GO:0018095) |
| 0.0 |
0.3 |
GO:0000712 |
resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 |
0.1 |
GO:0016561 |
protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.0 |
0.4 |
GO:0046688 |
response to copper ion(GO:0046688) |
| 0.0 |
0.1 |
GO:0042998 |
positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.0 |
0.1 |
GO:1904749 |
regulation of protein localization to nucleolus(GO:1904749) positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.0 |
0.1 |
GO:1903225 |
negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 |
0.1 |
GO:0048312 |
intracellular distribution of mitochondria(GO:0048312) |
| 0.0 |
0.3 |
GO:0007175 |
negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 |
0.2 |
GO:0030578 |
PML body organization(GO:0030578) |
| 0.0 |
0.1 |
GO:0000733 |
DNA strand renaturation(GO:0000733) |
| 0.0 |
0.4 |
GO:0042407 |
cristae formation(GO:0042407) |
| 0.0 |
0.1 |
GO:0034214 |
protein hexamerization(GO:0034214) |
| 0.0 |
0.5 |
GO:1902259 |
regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.0 |
0.3 |
GO:0006654 |
phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.0 |
0.1 |
GO:2000210 |
positive regulation of anoikis(GO:2000210) |
| 0.0 |
0.4 |
GO:0046597 |
negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 |
0.4 |
GO:0032438 |
melanosome organization(GO:0032438) anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 |
0.2 |
GO:0071372 |
cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.0 |
0.2 |
GO:0030322 |
stabilization of membrane potential(GO:0030322) |
| 0.0 |
0.2 |
GO:2000650 |
negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.0 |
0.3 |
GO:0010388 |
cullin deneddylation(GO:0010388) |
| 0.0 |
0.1 |
GO:1900127 |
positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 0.0 |
0.1 |
GO:0005981 |
regulation of glycogen catabolic process(GO:0005981) |
| 0.0 |
0.1 |
GO:0051013 |
microtubule severing(GO:0051013) |
| 0.0 |
0.1 |
GO:0002361 |
CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.0 |
0.4 |
GO:0008210 |
estrogen metabolic process(GO:0008210) |
| 0.0 |
0.5 |
GO:0022401 |
desensitization of G-protein coupled receptor protein signaling pathway(GO:0002029) negative adaptation of signaling pathway(GO:0022401) |
| 0.0 |
0.2 |
GO:0034983 |
peptidyl-lysine deacetylation(GO:0034983) |
| 0.0 |
0.1 |
GO:0002314 |
germinal center B cell differentiation(GO:0002314) |
| 0.0 |
0.5 |
GO:0015985 |
energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 |
0.1 |
GO:0009052 |
pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 |
0.2 |
GO:0042989 |
sequestering of actin monomers(GO:0042989) |
| 0.0 |
0.2 |
GO:0045187 |
regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.0 |
0.3 |
GO:0016558 |
protein import into peroxisome matrix(GO:0016558) |
| 0.0 |
0.8 |
GO:0022904 |
respiratory electron transport chain(GO:0022904) |
| 0.0 |
0.1 |
GO:2000467 |
positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.0 |
0.1 |
GO:0060715 |
syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.0 |
0.1 |
GO:2000786 |
positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 |
0.0 |
GO:0090202 |
transcriptional activation by promoter-enhancer looping(GO:0071733) regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.0 |
0.1 |
GO:0014886 |
transition between slow and fast fiber(GO:0014886) |
| 0.0 |
0.2 |
GO:0006309 |
apoptotic DNA fragmentation(GO:0006309) |
| 0.0 |
0.1 |
GO:0019433 |
triglyceride catabolic process(GO:0019433) |
| 0.0 |
0.1 |
GO:0072488 |
ammonium transmembrane transport(GO:0072488) |
| 0.0 |
0.1 |
GO:0006610 |
ribosomal protein import into nucleus(GO:0006610) |
| 0.0 |
0.0 |
GO:0039530 |
MDA-5 signaling pathway(GO:0039530) |
| 0.0 |
0.2 |
GO:0009083 |
branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 |
0.1 |
GO:1900112 |
regulation of histone H3-K9 trimethylation(GO:1900112) negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 |
0.5 |
GO:0090162 |
establishment of epithelial cell polarity(GO:0090162) |
| 0.0 |
0.1 |
GO:0036506 |
maintenance of unfolded protein(GO:0036506) protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.0 |
0.1 |
GO:0048752 |
semicircular canal morphogenesis(GO:0048752) |
| 0.0 |
0.2 |
GO:0006614 |
SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 |
0.2 |
GO:0001574 |
ganglioside biosynthetic process(GO:0001574) |
| 0.0 |
0.1 |
GO:0034551 |
respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 |
0.3 |
GO:0061014 |
positive regulation of mRNA catabolic process(GO:0061014) |
| 0.0 |
0.9 |
GO:0035914 |
skeletal muscle cell differentiation(GO:0035914) |
| 0.0 |
0.1 |
GO:0006390 |
transcription from mitochondrial promoter(GO:0006390) |
| 0.0 |
0.1 |
GO:0090084 |
negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 |
0.1 |
GO:0036444 |
calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 |
0.2 |
GO:0010614 |
negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 |
0.2 |
GO:0071157 |
negative regulation of cell cycle arrest(GO:0071157) |