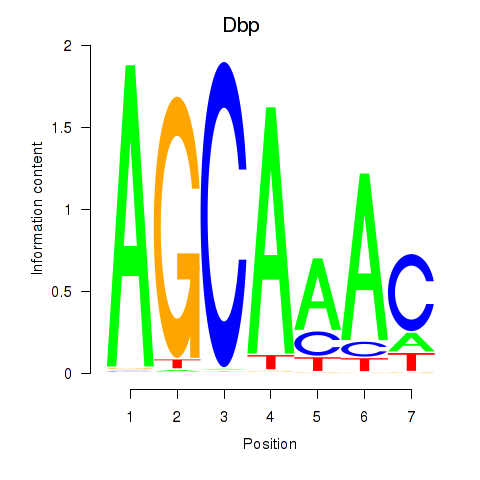
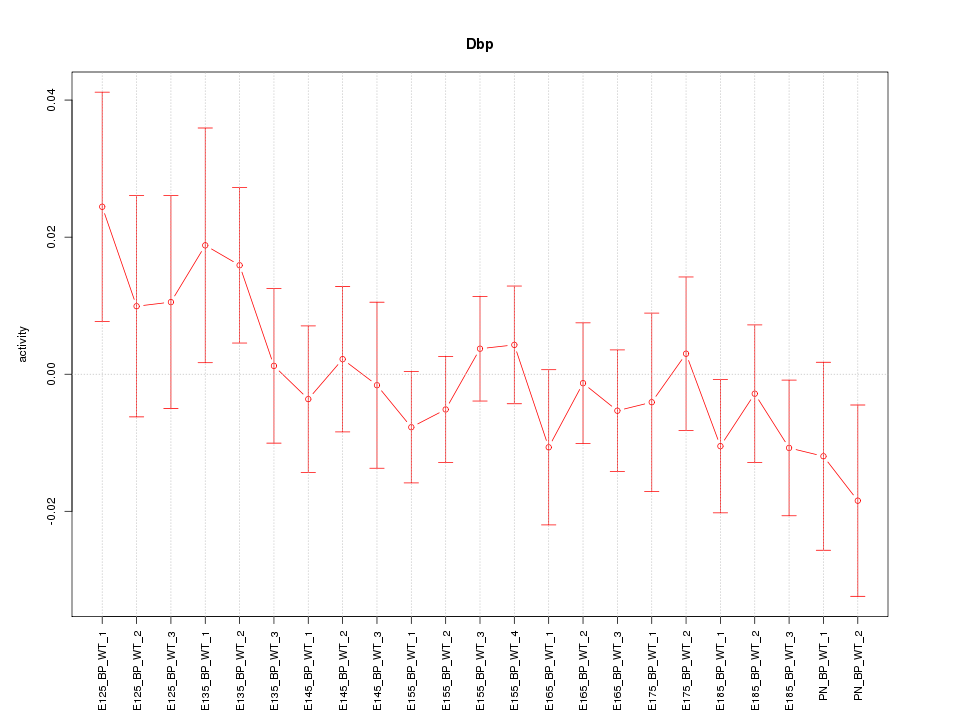
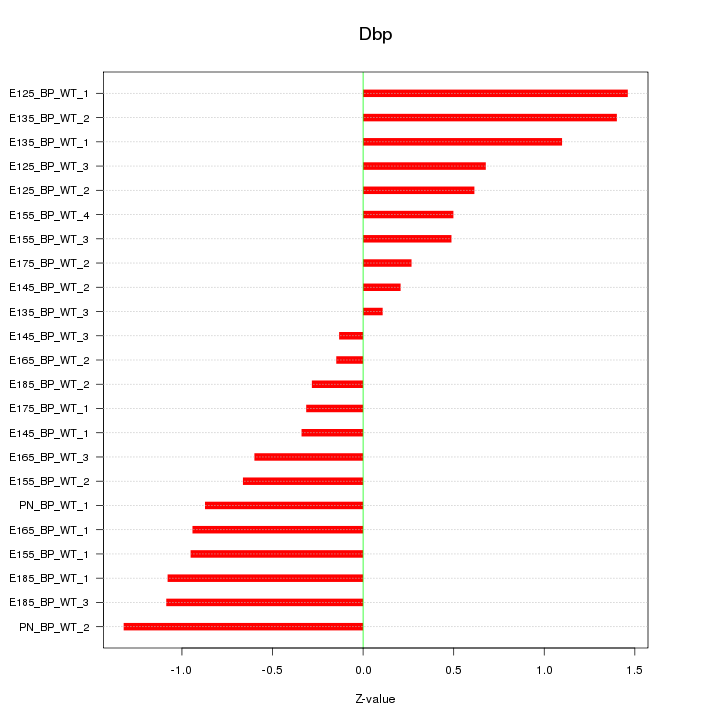
| 1.3 |
7.8 |
GO:0021797 |
forebrain anterior/posterior pattern specification(GO:0021797) |
| 1.1 |
3.2 |
GO:0003345 |
proepicardium cell migration involved in pericardium morphogenesis(GO:0003345) |
| 1.0 |
5.7 |
GO:0060024 |
rhythmic synaptic transmission(GO:0060024) |
| 0.7 |
2.0 |
GO:0090258 |
negative regulation of mitochondrial fission(GO:0090258) |
| 0.5 |
1.5 |
GO:0016115 |
terpenoid catabolic process(GO:0016115) |
| 0.4 |
1.2 |
GO:0060023 |
soft palate development(GO:0060023) |
| 0.4 |
1.9 |
GO:0030644 |
cellular chloride ion homeostasis(GO:0030644) |
| 0.3 |
0.6 |
GO:0051968 |
positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.3 |
1.8 |
GO:0061052 |
negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.3 |
0.9 |
GO:0097155 |
fasciculation of sensory neuron axon(GO:0097155) |
| 0.3 |
0.9 |
GO:2000569 |
T-helper 2 cell activation(GO:0035712) positive regulation of T-helper 17 type immune response(GO:2000318) regulation of T-helper 2 cell activation(GO:2000569) positive regulation of T-helper 2 cell activation(GO:2000570) |
| 0.3 |
0.8 |
GO:0032474 |
otolith morphogenesis(GO:0032474) |
| 0.3 |
1.3 |
GO:0051661 |
maintenance of centrosome location(GO:0051661) |
| 0.2 |
2.0 |
GO:0021937 |
cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) |
| 0.2 |
1.7 |
GO:0071361 |
cellular response to zinc ion(GO:0071294) cellular response to ethanol(GO:0071361) |
| 0.2 |
2.8 |
GO:0046643 |
regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.2 |
2.4 |
GO:0060501 |
positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.2 |
2.0 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.2 |
2.1 |
GO:1900383 |
regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.2 |
2.5 |
GO:1902993 |
positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.2 |
0.6 |
GO:0071930 |
negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) lens fiber cell apoptotic process(GO:1990086) |
| 0.2 |
0.3 |
GO:0034372 |
very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.2 |
0.8 |
GO:0010792 |
DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.2 |
0.8 |
GO:0040032 |
post-embryonic body morphogenesis(GO:0040032) |
| 0.1 |
1.5 |
GO:0014824 |
artery smooth muscle contraction(GO:0014824) |
| 0.1 |
0.4 |
GO:0001928 |
regulation of exocyst assembly(GO:0001928) |
| 0.1 |
1.6 |
GO:0048672 |
positive regulation of collateral sprouting(GO:0048672) |
| 0.1 |
1.1 |
GO:0090043 |
regulation of tubulin deacetylation(GO:0090043) |
| 0.1 |
0.6 |
GO:0010993 |
regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.1 |
0.4 |
GO:0048143 |
astrocyte activation(GO:0048143) |
| 0.1 |
3.1 |
GO:0007214 |
gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 |
0.4 |
GO:0061300 |
cerebellum vasculature development(GO:0061300) |
| 0.1 |
2.9 |
GO:0010569 |
regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.1 |
0.5 |
GO:0033128 |
negative regulation of histone phosphorylation(GO:0033128) positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.1 |
0.6 |
GO:0006344 |
maintenance of chromatin silencing(GO:0006344) |
| 0.1 |
0.7 |
GO:0006287 |
base-excision repair, gap-filling(GO:0006287) |
| 0.1 |
0.6 |
GO:0051694 |
pointed-end actin filament capping(GO:0051694) |
| 0.1 |
0.2 |
GO:0032877 |
positive regulation of DNA endoreduplication(GO:0032877) |
| 0.1 |
1.2 |
GO:0006610 |
ribosomal protein import into nucleus(GO:0006610) |
| 0.1 |
0.5 |
GO:0036324 |
vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.1 |
0.6 |
GO:0045003 |
DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.1 |
0.4 |
GO:0035331 |
negative regulation of hippo signaling(GO:0035331) |
| 0.1 |
0.3 |
GO:0006553 |
lysine metabolic process(GO:0006553) |
| 0.1 |
0.3 |
GO:0007182 |
common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.1 |
0.2 |
GO:0021612 |
facial nerve structural organization(GO:0021612) |
| 0.1 |
0.5 |
GO:1900028 |
negative regulation of ruffle assembly(GO:1900028) |
| 0.1 |
0.2 |
GO:2000224 |
testosterone biosynthetic process(GO:0061370) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.1 |
0.2 |
GO:0061386 |
closure of optic fissure(GO:0061386) |
| 0.1 |
0.4 |
GO:1900186 |
negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 0.1 |
0.5 |
GO:0007258 |
JUN phosphorylation(GO:0007258) |
| 0.1 |
0.6 |
GO:0043353 |
enucleate erythrocyte differentiation(GO:0043353) |
| 0.1 |
0.8 |
GO:0019065 |
receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.1 |
0.6 |
GO:0032625 |
interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 |
5.1 |
GO:0051384 |
response to glucocorticoid(GO:0051384) |
| 0.1 |
0.3 |
GO:0070934 |
CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 |
0.6 |
GO:0061368 |
behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.1 |
0.4 |
GO:2000623 |
regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 |
0.7 |
GO:0030007 |
cellular potassium ion homeostasis(GO:0030007) |
| 0.1 |
1.8 |
GO:0003301 |
physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 |
0.2 |
GO:1900158 |
negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.1 |
0.6 |
GO:0061158 |
3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.1 |
0.9 |
GO:0015937 |
coenzyme A biosynthetic process(GO:0015937) |
| 0.1 |
1.4 |
GO:0006359 |
regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.1 |
4.6 |
GO:0010923 |
negative regulation of phosphatase activity(GO:0010923) |
| 0.1 |
0.7 |
GO:2000741 |
positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.1 |
4.0 |
GO:0060395 |
SMAD protein signal transduction(GO:0060395) |
| 0.1 |
0.2 |
GO:0046294 |
formaldehyde catabolic process(GO:0046294) |
| 0.1 |
0.6 |
GO:0021559 |
trigeminal nerve development(GO:0021559) |
| 0.1 |
0.1 |
GO:0034310 |
primary alcohol catabolic process(GO:0034310) |
| 0.1 |
0.3 |
GO:1901029 |
negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.1 |
0.7 |
GO:0070986 |
left/right axis specification(GO:0070986) |
| 0.1 |
0.2 |
GO:0002408 |
myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.1 |
2.5 |
GO:0030901 |
midbrain development(GO:0030901) |
| 0.0 |
0.4 |
GO:0000083 |
regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 |
2.0 |
GO:0048013 |
ephrin receptor signaling pathway(GO:0048013) |
| 0.0 |
0.3 |
GO:0089711 |
L-glutamate transmembrane transport(GO:0089711) |
| 0.0 |
0.6 |
GO:0060837 |
blood vessel endothelial cell differentiation(GO:0060837) |
| 0.0 |
0.5 |
GO:1902018 |
negative regulation of cilium assembly(GO:1902018) |
| 0.0 |
0.3 |
GO:0035735 |
intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 |
0.2 |
GO:0006177 |
GMP biosynthetic process(GO:0006177) |
| 0.0 |
0.6 |
GO:2000811 |
negative regulation of anoikis(GO:2000811) |
| 0.0 |
0.5 |
GO:0048146 |
positive regulation of fibroblast proliferation(GO:0048146) |
| 0.0 |
0.9 |
GO:0043171 |
peptide catabolic process(GO:0043171) |
| 0.0 |
0.4 |
GO:0009312 |
oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 |
0.1 |
GO:0048014 |
Tie signaling pathway(GO:0048014) |
| 0.0 |
0.6 |
GO:0010669 |
epithelial structure maintenance(GO:0010669) |
| 0.0 |
0.5 |
GO:0010603 |
regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 |
0.5 |
GO:0071218 |
cellular response to misfolded protein(GO:0071218) |
| 0.0 |
0.4 |
GO:0030574 |
collagen catabolic process(GO:0030574) |
| 0.0 |
0.2 |
GO:0051639 |
actin filament network formation(GO:0051639) |
| 0.0 |
0.6 |
GO:0071425 |
hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 |
0.6 |
GO:0010842 |
retina layer formation(GO:0010842) |
| 0.0 |
1.9 |
GO:0001578 |
microtubule bundle formation(GO:0001578) |
| 0.0 |
1.2 |
GO:0007200 |
phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.0 |
0.3 |
GO:0002115 |
store-operated calcium entry(GO:0002115) |
| 0.0 |
0.2 |
GO:0070842 |
aggresome assembly(GO:0070842) |
| 0.0 |
0.6 |
GO:0048741 |
skeletal muscle fiber development(GO:0048741) |
| 0.0 |
0.3 |
GO:0009263 |
deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 |
0.3 |
GO:0034142 |
toll-like receptor 4 signaling pathway(GO:0034142) |
| 0.0 |
0.6 |
GO:1902476 |
chloride transmembrane transport(GO:1902476) |
| 0.0 |
0.1 |
GO:0043312 |
neutrophil degranulation(GO:0043312) |
| 0.0 |
0.4 |
GO:0043984 |
histone H4-K16 acetylation(GO:0043984) |
| 0.0 |
0.2 |
GO:1900364 |
regulation of mRNA polyadenylation(GO:1900363) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 |
1.0 |
GO:0061077 |
chaperone-mediated protein folding(GO:0061077) |
| 0.0 |
0.4 |
GO:0071625 |
vocalization behavior(GO:0071625) |
| 0.0 |
0.8 |
GO:1904893 |
negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 |
0.2 |
GO:0021527 |
spinal cord association neuron differentiation(GO:0021527) |
| 0.0 |
0.3 |
GO:0055021 |
regulation of cardiac muscle tissue growth(GO:0055021) |
| 0.0 |
0.2 |
GO:0008090 |
retrograde axonal transport(GO:0008090) |
| 0.0 |
0.3 |
GO:0007020 |
microtubule nucleation(GO:0007020) |
| 0.0 |
0.0 |
GO:0032899 |
regulation of neurotrophin production(GO:0032899) |
| 0.0 |
0.4 |
GO:0045739 |
positive regulation of DNA repair(GO:0045739) |
| 0.0 |
0.2 |
GO:0048026 |
positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 |
0.1 |
GO:0097368 |
establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 |
0.1 |
GO:0006452 |
translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |