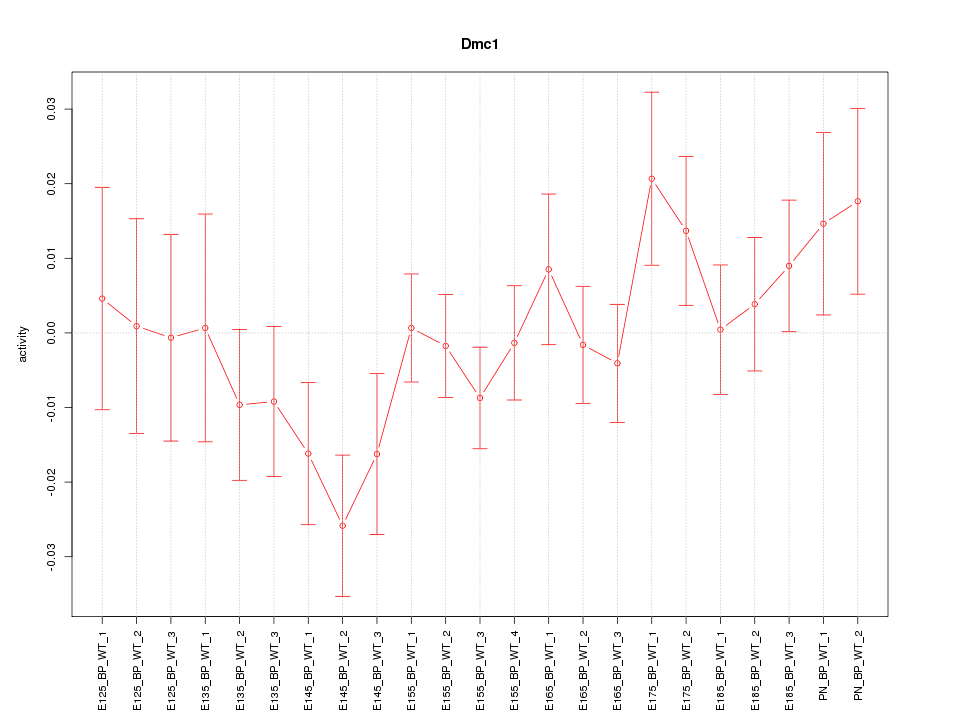
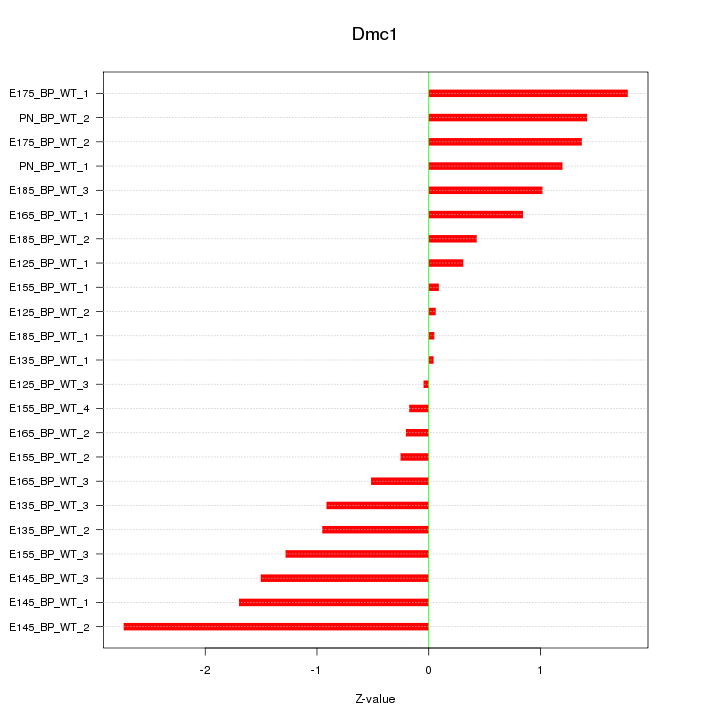
Motif ID: Dmc1
Z-value: 1.082
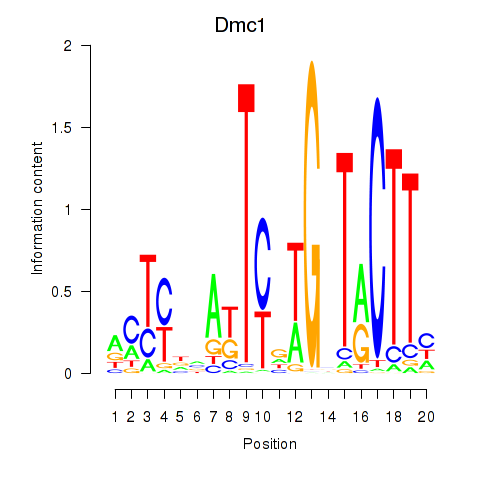
Transcription factors associated with Dmc1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Dmc1 | ENSMUSG00000022429.10 | Dmc1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Dmc1 | mm10_v2_chr15_-_79605084_79605114 | 0.52 | 1.1e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0051464 | positive regulation of cortisol secretion(GO:0051464) |
| 0.6 | 1.8 | GO:0008228 | opsonization(GO:0008228) modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.6 | 1.7 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.4 | 2.2 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.4 | 1.8 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.3 | 1.0 | GO:0032632 | interleukin-3 production(GO:0032632) |
| 0.3 | 1.2 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.3 | 1.8 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.3 | 2.5 | GO:0042447 | hormone catabolic process(GO:0042447) |
| 0.3 | 0.8 | GO:0032240 | RNA import into nucleus(GO:0006404) mRNA export from nucleus in response to heat stress(GO:0031990) negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.2 | 1.4 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.2 | 0.9 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.2 | 1.1 | GO:2000425 | regulation of apoptotic cell clearance(GO:2000425) |
| 0.2 | 1.2 | GO:1903887 | motile primary cilium assembly(GO:1903887) protein localization to ciliary transition zone(GO:1904491) |
| 0.2 | 0.6 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.2 | 0.4 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.2 | 0.6 | GO:1903795 | regulation of inorganic anion transmembrane transport(GO:1903795) |
| 0.2 | 0.6 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.2 | 1.3 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.2 | 0.9 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) |
| 0.2 | 0.5 | GO:0042196 | dichloromethane metabolic process(GO:0018900) chlorinated hydrocarbon metabolic process(GO:0042196) halogenated hydrocarbon metabolic process(GO:0042197) |
| 0.2 | 1.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.2 | 0.5 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.2 | 0.7 | GO:0000019 | regulation of mitotic recombination(GO:0000019) |
| 0.2 | 0.8 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.2 | 1.0 | GO:0032229 | positive regulation of gamma-aminobutyric acid secretion(GO:0014054) negative regulation of synaptic transmission, GABAergic(GO:0032229) |
| 0.2 | 0.8 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.2 | 0.8 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.2 | 0.6 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.1 | 1.5 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.1 | 0.7 | GO:2001170 | negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.1 | 1.3 | GO:0006544 | glycine metabolic process(GO:0006544) |
| 0.1 | 0.5 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.1 | 0.9 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.1 | 0.6 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.1 | 1.1 | GO:0043206 | extracellular fibril organization(GO:0043206) |
| 0.1 | 0.6 | GO:0002553 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.1 | 0.3 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.1 | 0.4 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.1 | 0.3 | GO:0034241 | positive regulation of macrophage fusion(GO:0034241) regulation of osteoclast proliferation(GO:0090289) |
| 0.1 | 0.2 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.1 | 0.4 | GO:1903416 | negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) regulation of calcidiol 1-monooxygenase activity(GO:0060558) response to glycoside(GO:1903416) |
| 0.1 | 1.7 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.1 | 0.3 | GO:2000819 | regulation of nucleotide-excision repair(GO:2000819) |
| 0.1 | 0.3 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.1 | 2.7 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 1.8 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.1 | 0.5 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.1 | 0.7 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.1 | 0.5 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.1 | 0.8 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 0.4 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 0.7 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.1 | 1.2 | GO:0018126 | protein hydroxylation(GO:0018126) |
| 0.1 | 0.2 | GO:2001187 | positive regulation of CD8-positive, alpha-beta T cell activation(GO:2001187) |
| 0.1 | 0.7 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.1 | 0.6 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.1 | 0.2 | GO:0046077 | dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 0.1 | 0.6 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.1 | 0.7 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 | 0.3 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.1 | 0.3 | GO:0051593 | response to folic acid(GO:0051593) |
| 0.1 | 0.1 | GO:0048793 | pronephros development(GO:0048793) |
| 0.1 | 0.3 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.1 | 0.2 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.1 | 0.1 | GO:0002371 | dendritic cell cytokine production(GO:0002371) regulation of dendritic cell cytokine production(GO:0002730) |
| 0.1 | 1.9 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.1 | 0.3 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.1 | 0.9 | GO:0046519 | sphingoid metabolic process(GO:0046519) |
| 0.1 | 1.3 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.1 | 0.6 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.1 | 0.5 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.1 | 0.5 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.1 | 0.3 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.1 | 0.3 | GO:0050857 | positive regulation of antigen receptor-mediated signaling pathway(GO:0050857) |
| 0.1 | 0.2 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.1 | 0.3 | GO:0042148 | strand invasion(GO:0042148) |
| 0.1 | 0.7 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 0.5 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.1 | 0.2 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) negative regulation of negative chemotaxis(GO:0050925) |
| 0.0 | 0.3 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.3 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.5 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.1 | GO:0002339 | B cell selection(GO:0002339) B cell negative selection(GO:0002352) establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.0 | 0.4 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.0 | 0.1 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.0 | 0.3 | GO:0042045 | epithelial fluid transport(GO:0042045) |
| 0.0 | 2.4 | GO:0009063 | cellular amino acid catabolic process(GO:0009063) |
| 0.0 | 0.8 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.5 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.0 | 0.2 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.0 | 0.5 | GO:0001711 | endodermal cell fate commitment(GO:0001711) |
| 0.0 | 0.7 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 0.7 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.0 | 0.3 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.5 | GO:0071467 | cellular response to pH(GO:0071467) |
| 0.0 | 0.6 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.2 | GO:0048266 | response to pain(GO:0048265) behavioral response to pain(GO:0048266) |
| 0.0 | 0.5 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 1.2 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.1 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.0 | 0.8 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 0.3 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.4 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.5 | GO:0034308 | primary alcohol metabolic process(GO:0034308) |
| 0.0 | 0.5 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.3 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.0 | 0.1 | GO:0006097 | glyoxylate cycle(GO:0006097) glyoxylate metabolic process(GO:0046487) |
| 0.0 | 0.4 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.1 | GO:0055071 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.0 | 0.6 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 1.0 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.0 | 0.2 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.1 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.0 | GO:0048597 | post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.0 | 0.4 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.0 | 0.4 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.1 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.1 | GO:0021984 | adenohypophysis development(GO:0021984) |
| 0.0 | 1.8 | GO:0098780 | mitophagy in response to mitochondrial depolarization(GO:0098779) response to mitochondrial depolarisation(GO:0098780) |
| 0.0 | 0.5 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.6 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.6 | GO:0006360 | transcription from RNA polymerase I promoter(GO:0006360) |
| 0.0 | 0.1 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 | 0.1 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 | 0.4 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.0 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.5 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.2 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.4 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.4 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.8 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.0 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.5 | 1.5 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.2 | 0.7 | GO:1903095 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 0.2 | 1.0 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.2 | 0.6 | GO:0034684 | integrin alphav-beta5 complex(GO:0034684) |
| 0.2 | 1.3 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.2 | 0.6 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.2 | 0.5 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.2 | 0.8 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.1 | 0.6 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.1 | 0.9 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.1 | 1.5 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 0.5 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.1 | 1.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 0.7 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 0.9 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.1 | 0.3 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.1 | 1.1 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.1 | 1.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 1.8 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.1 | 0.4 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 0.4 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.1 | 1.0 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.1 | 0.5 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 0.9 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.1 | 0.4 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 0.6 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.1 | 1.6 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.4 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 2.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.3 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.5 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.5 | GO:0005675 | DNA replication factor A complex(GO:0005662) holo TFIIH complex(GO:0005675) |
| 0.0 | 0.2 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.3 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.3 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.3 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.7 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 0.7 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.1 | GO:0044299 | C-fiber(GO:0044299) |
| 0.0 | 0.7 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.1 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 0.8 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.4 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.3 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.2 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.4 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.0 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.6 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.4 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.1 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 0.1 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.4 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.5 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.9 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.9 | 2.6 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.6 | 1.8 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.4 | 1.3 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.4 | 1.8 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.4 | 2.5 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.3 | 1.3 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.3 | 1.0 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.3 | 1.2 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.3 | 1.8 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.2 | 0.6 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.2 | 0.6 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.2 | 1.8 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.2 | 0.5 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.2 | 0.7 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.2 | 1.3 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.2 | 1.2 | GO:0016918 | retinal binding(GO:0016918) |
| 0.2 | 0.5 | GO:0019120 | hydrolase activity, acting on acid halide bonds(GO:0016824) hydrolase activity, acting on acid halide bonds, in C-halide compounds(GO:0019120) alkylhalidase activity(GO:0047651) |
| 0.2 | 0.7 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.2 | 0.8 | GO:0018121 | imidazoleglycerol-phosphate synthase activity(GO:0000107) NAD(P)-cysteine ADP-ribosyltransferase activity(GO:0018071) NAD(P)-asparagine ADP-ribosyltransferase activity(GO:0018121) NAD(P)-serine ADP-ribosyltransferase activity(GO:0018127) 7-cyano-7-deazaguanine tRNA-ribosyltransferase activity(GO:0043867) purine deoxyribosyltransferase activity(GO:0044102) |
| 0.2 | 1.6 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.2 | 0.5 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.2 | 0.8 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 0.4 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) |
| 0.1 | 0.4 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.1 | 0.5 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.1 | 0.3 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.1 | 0.3 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) single guanine insertion binding(GO:0032142) |
| 0.1 | 0.3 | GO:0043912 | D-lysine oxidase activity(GO:0043912) |
| 0.1 | 2.7 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) |
| 0.1 | 0.4 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.4 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 0.9 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.1 | 0.5 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.1 | 0.5 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.1 | 0.6 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.4 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.1 | 1.5 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) |
| 0.1 | 1.8 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.1 | 1.0 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.1 | 0.4 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.1 | 0.6 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 0.3 | GO:0004104 | cholinesterase activity(GO:0004104) choline binding(GO:0033265) |
| 0.1 | 0.2 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.1 | 0.7 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.1 | 0.5 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.1 | 0.3 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.1 | 0.2 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.1 | 0.3 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 0.2 | GO:0019863 | IgE binding(GO:0019863) |
| 0.1 | 0.5 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.1 | 0.3 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 0.8 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.3 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 0.3 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 0.2 | GO:0004954 | icosanoid receptor activity(GO:0004953) prostanoid receptor activity(GO:0004954) prostaglandin receptor activity(GO:0004955) |
| 0.1 | 0.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.5 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.1 | 0.3 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.1 | 1.7 | GO:0034930 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) heparan sulfate 2-O-sulfotransferase activity(GO:0004394) HNK-1 sulfotransferase activity(GO:0016232) heparan sulfate 6-O-sulfotransferase activity(GO:0017095) trans-9R,10R-dihydrodiolphenanthrene sulfotransferase activity(GO:0018721) 1-phenanthrol sulfotransferase activity(GO:0018722) 3-phenanthrol sulfotransferase activity(GO:0018723) 4-phenanthrol sulfotransferase activity(GO:0018724) trans-3,4-dihydrodiolphenanthrene sulfotransferase activity(GO:0018725) 9-phenanthrol sulfotransferase activity(GO:0018726) 2-phenanthrol sulfotransferase activity(GO:0018727) phenanthrol sulfotransferase activity(GO:0019111) 1-hydroxypyrene sulfotransferase activity(GO:0034930) proteoglycan sulfotransferase activity(GO:0050698) cholesterol sulfotransferase activity(GO:0051922) hydroxyjasmonate sulfotransferase activity(GO:0080131) |
| 0.1 | 0.3 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.1 | 0.5 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.6 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.3 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 1.3 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.3 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 1.0 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.8 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.5 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.2 | GO:0050347 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.0 | 0.3 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 0.3 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 3.0 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.6 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 4.0 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.2 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.1 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 0.4 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.1 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.0 | 0.1 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.0 | 0.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.1 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.0 | 1.1 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 0.4 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.3 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 1.0 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.8 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.2 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.0 | 0.8 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.5 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.7 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.0 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 0.9 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 1.6 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.8 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.2 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.1 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.5 | GO:0044824 | integrase activity(GO:0008907) T/G mismatch-specific endonuclease activity(GO:0043765) retroviral integrase activity(GO:0044823) retroviral 3' processing activity(GO:0044824) |
| 0.0 | 0.7 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.9 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 2.1 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.9 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 1.6 | GO:0016829 | lyase activity(GO:0016829) |