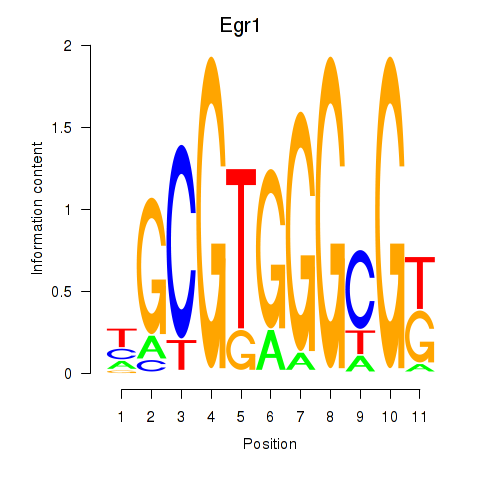
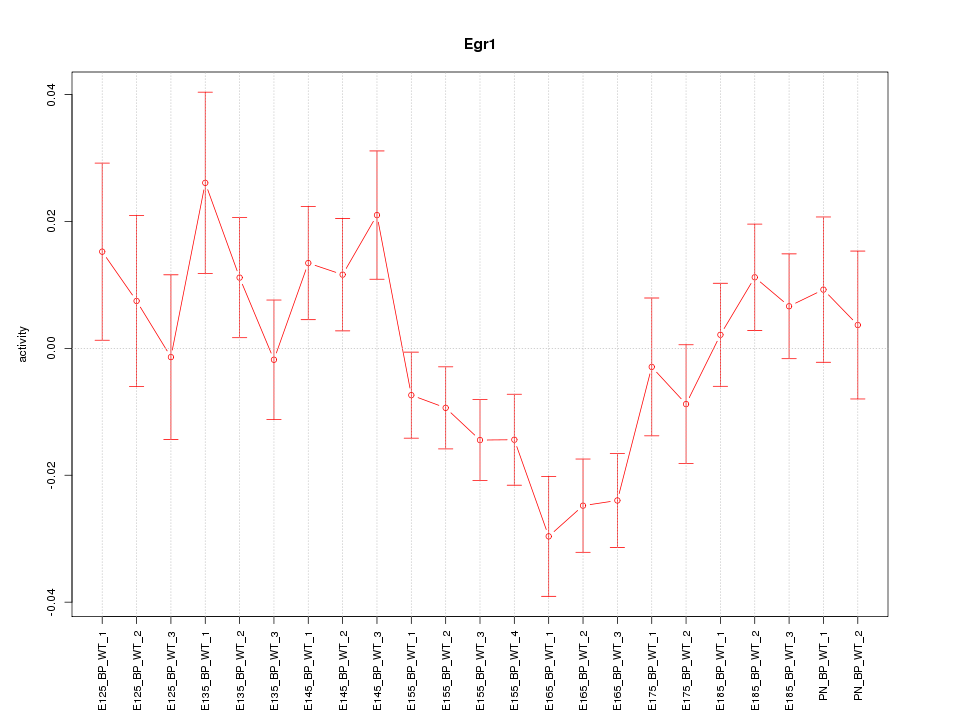
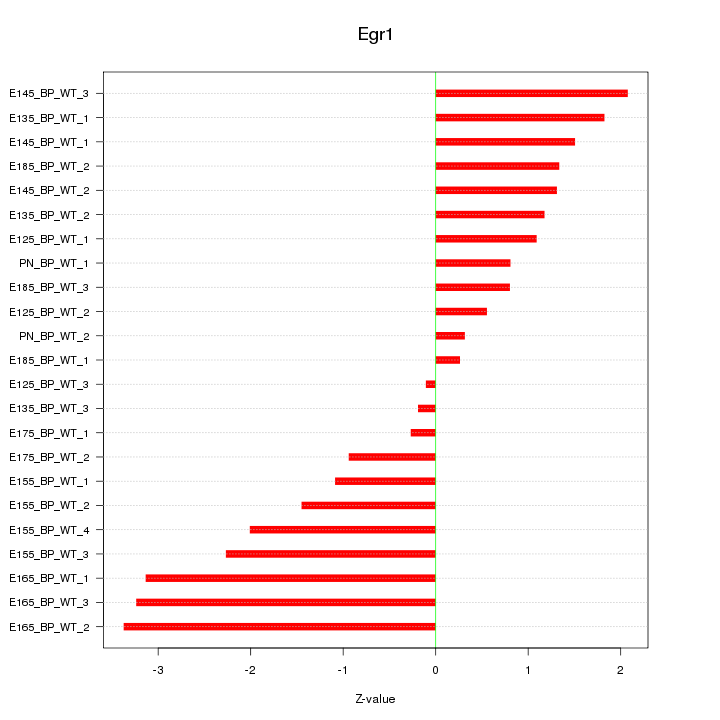
| 2.6 |
13.1 |
GO:2000325 |
regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 1.1 |
3.4 |
GO:0007621 |
negative regulation of female receptivity(GO:0007621) |
| 0.7 |
2.2 |
GO:0030070 |
insulin processing(GO:0030070) |
| 0.7 |
2.1 |
GO:0086017 |
response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) Purkinje myocyte action potential(GO:0086017) |
| 0.7 |
4.2 |
GO:0032796 |
uropod organization(GO:0032796) |
| 0.6 |
1.9 |
GO:0010752 |
negative regulation of antigen processing and presentation(GO:0002578) negative regulation of nitric oxide mediated signal transduction(GO:0010751) regulation of cGMP-mediated signaling(GO:0010752) negative regulation of plasminogen activation(GO:0010757) |
| 0.6 |
1.8 |
GO:0060010 |
Sertoli cell fate commitment(GO:0060010) |
| 0.5 |
2.2 |
GO:0002457 |
T cell antigen processing and presentation(GO:0002457) |
| 0.5 |
1.5 |
GO:0010512 |
negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) |
| 0.5 |
0.5 |
GO:0071374 |
cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.4 |
1.3 |
GO:0046381 |
CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 0.4 |
1.6 |
GO:0010040 |
response to iron(II) ion(GO:0010040) positive regulation of hydrogen peroxide metabolic process(GO:0010726) negative regulation of synaptic transmission, dopaminergic(GO:0032227) negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) cellular response to copper ion(GO:0071280) regulation of peroxidase activity(GO:2000468) |
| 0.4 |
1.9 |
GO:1902683 |
regulation of receptor localization to synapse(GO:1902683) |
| 0.4 |
1.1 |
GO:0060598 |
dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 0.4 |
4.2 |
GO:0051967 |
negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.4 |
0.4 |
GO:0086068 |
Purkinje myocyte to ventricular cardiac muscle cell signaling(GO:0086029) Purkinje myocyte to ventricular cardiac muscle cell communication(GO:0086068) |
| 0.4 |
1.1 |
GO:1900673 |
olefin metabolic process(GO:1900673) |
| 0.4 |
2.5 |
GO:0035902 |
response to immobilization stress(GO:0035902) |
| 0.3 |
0.9 |
GO:0048211 |
Golgi vesicle docking(GO:0048211) |
| 0.3 |
1.8 |
GO:0097491 |
trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.3 |
1.4 |
GO:1902031 |
regulation of NADP metabolic process(GO:1902031) |
| 0.3 |
1.9 |
GO:0016480 |
negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.3 |
2.2 |
GO:1903423 |
positive regulation of synaptic vesicle recycling(GO:1903423) |
| 0.3 |
1.1 |
GO:0060741 |
prostate gland stromal morphogenesis(GO:0060741) |
| 0.3 |
0.8 |
GO:0050883 |
musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.3 |
2.4 |
GO:0010587 |
miRNA catabolic process(GO:0010587) |
| 0.2 |
0.7 |
GO:0060437 |
lung growth(GO:0060437) |
| 0.2 |
1.5 |
GO:1900028 |
negative regulation of ruffle assembly(GO:1900028) |
| 0.2 |
1.0 |
GO:2000313 |
fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.2 |
1.7 |
GO:1900107 |
regulation of nodal signaling pathway(GO:1900107) |
| 0.2 |
0.7 |
GO:0033634 |
positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.2 |
1.1 |
GO:0034436 |
glycoprotein transport(GO:0034436) |
| 0.2 |
1.6 |
GO:2000124 |
regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.2 |
0.9 |
GO:2000304 |
positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.2 |
1.3 |
GO:1900451 |
positive regulation of glutamate receptor signaling pathway(GO:1900451) positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.2 |
1.5 |
GO:1900116 |
sequestering of extracellular ligand from receptor(GO:0035581) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.2 |
0.8 |
GO:0070086 |
ubiquitin-dependent endocytosis(GO:0070086) |
| 0.2 |
1.2 |
GO:0045162 |
clustering of voltage-gated sodium channels(GO:0045162) |
| 0.2 |
0.6 |
GO:0045819 |
positive regulation of glycogen catabolic process(GO:0045819) |
| 0.2 |
0.6 |
GO:0035519 |
protein K29-linked ubiquitination(GO:0035519) |
| 0.2 |
4.0 |
GO:0032897 |
negative regulation of viral transcription(GO:0032897) |
| 0.2 |
0.6 |
GO:0006597 |
spermine biosynthetic process(GO:0006597) |
| 0.2 |
1.3 |
GO:0001771 |
immunological synapse formation(GO:0001771) |
| 0.2 |
0.5 |
GO:0021849 |
neuroblast division in subventricular zone(GO:0021849) |
| 0.2 |
1.0 |
GO:0015015 |
heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.2 |
7.1 |
GO:0007189 |
adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.2 |
2.9 |
GO:2000311 |
regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.2 |
1.0 |
GO:0019695 |
choline metabolic process(GO:0019695) |
| 0.1 |
1.2 |
GO:0070317 |
negative regulation of G0 to G1 transition(GO:0070317) |
| 0.1 |
1.5 |
GO:0033008 |
positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.1 |
0.1 |
GO:0071677 |
positive regulation of mononuclear cell migration(GO:0071677) |
| 0.1 |
0.4 |
GO:0033159 |
negative regulation of protein import into nucleus, translocation(GO:0033159) protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 |
0.7 |
GO:1902309 |
negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.1 |
2.5 |
GO:0051988 |
regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.1 |
0.8 |
GO:0006452 |
translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 |
0.4 |
GO:0001869 |
regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.1 |
0.4 |
GO:0070172 |
positive regulation of tooth mineralization(GO:0070172) |
| 0.1 |
0.4 |
GO:1903244 |
positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.1 |
0.9 |
GO:2000192 |
negative regulation of fatty acid transport(GO:2000192) |
| 0.1 |
0.3 |
GO:0070634 |
transepithelial ammonium transport(GO:0070634) |
| 0.1 |
2.1 |
GO:0031340 |
positive regulation of vesicle fusion(GO:0031340) |
| 0.1 |
1.8 |
GO:1900454 |
positive regulation of long term synaptic depression(GO:1900454) |
| 0.1 |
0.2 |
GO:0021529 |
spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.1 |
0.4 |
GO:0039536 |
negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.1 |
0.9 |
GO:0060120 |
auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.1 |
0.4 |
GO:0009597 |
detection of virus(GO:0009597) |
| 0.1 |
0.5 |
GO:1902570 |
protein localization to nucleolus(GO:1902570) |
| 0.1 |
0.3 |
GO:1902109 |
negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.1 |
3.6 |
GO:0045746 |
negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 |
0.5 |
GO:1900016 |
negative regulation of cytokine production involved in inflammatory response(GO:1900016) |
| 0.1 |
1.3 |
GO:2000480 |
negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 |
0.3 |
GO:0034392 |
negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.1 |
1.5 |
GO:0051764 |
actin crosslink formation(GO:0051764) |
| 0.1 |
0.4 |
GO:0002949 |
tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 |
0.5 |
GO:0071947 |
protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.1 |
0.7 |
GO:1903715 |
regulation of aerobic respiration(GO:1903715) |
| 0.1 |
2.4 |
GO:0035855 |
megakaryocyte development(GO:0035855) |
| 0.1 |
0.7 |
GO:0016584 |
nucleosome positioning(GO:0016584) |
| 0.1 |
1.0 |
GO:0006048 |
UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.1 |
0.3 |
GO:0051660 |
cortical microtubule organization(GO:0043622) establishment of centrosome localization(GO:0051660) |
| 0.1 |
0.5 |
GO:0006543 |
glutamine catabolic process(GO:0006543) |
| 0.1 |
0.7 |
GO:0034047 |
regulation of protein phosphatase type 2A activity(GO:0034047) |
| 0.1 |
0.6 |
GO:0090267 |
positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.1 |
0.5 |
GO:0007253 |
cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.1 |
0.2 |
GO:1903960 |
negative regulation of anion transmembrane transport(GO:1903960) |
| 0.1 |
0.4 |
GO:0030576 |
Cajal body organization(GO:0030576) |
| 0.1 |
0.3 |
GO:0006106 |
fumarate metabolic process(GO:0006106) aspartate catabolic process(GO:0006533) alditol biosynthetic process(GO:0019401) |
| 0.1 |
0.3 |
GO:1900126 |
negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.1 |
0.3 |
GO:0031509 |
telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.1 |
2.9 |
GO:0006879 |
cellular iron ion homeostasis(GO:0006879) |
| 0.1 |
1.0 |
GO:0051481 |
negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.1 |
0.6 |
GO:0045723 |
positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.1 |
0.4 |
GO:0018125 |
peptidyl-cysteine methylation(GO:0018125) |
| 0.1 |
0.1 |
GO:0007521 |
muscle cell fate determination(GO:0007521) |
| 0.1 |
0.6 |
GO:2000074 |
regulation of type B pancreatic cell development(GO:2000074) |
| 0.1 |
3.1 |
GO:0034605 |
cellular response to heat(GO:0034605) |
| 0.1 |
1.9 |
GO:0031065 |
positive regulation of histone deacetylation(GO:0031065) |
| 0.1 |
0.4 |
GO:2000507 |
positive regulation of energy homeostasis(GO:2000507) |
| 0.1 |
0.6 |
GO:2000601 |
positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 |
0.4 |
GO:0010944 |
negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.1 |
0.2 |
GO:1903334 |
positive regulation of protein folding(GO:1903334) |
| 0.1 |
0.8 |
GO:2001135 |
regulation of endocytic recycling(GO:2001135) |
| 0.1 |
3.5 |
GO:0045682 |
regulation of epidermis development(GO:0045682) |
| 0.1 |
0.2 |
GO:0045656 |
negative regulation of monocyte differentiation(GO:0045656) |
| 0.1 |
0.3 |
GO:0032485 |
regulation of Ral protein signal transduction(GO:0032485) |
| 0.1 |
1.0 |
GO:0048387 |
negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 |
0.3 |
GO:0003376 |
sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.1 |
1.9 |
GO:0070979 |
protein K11-linked ubiquitination(GO:0070979) |
| 0.1 |
0.6 |
GO:0033574 |
response to testosterone(GO:0033574) |
| 0.1 |
0.1 |
GO:1902498 |
regulation of protein autoubiquitination(GO:1902498) |
| 0.1 |
0.4 |
GO:0031119 |
tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 |
0.8 |
GO:0032927 |
positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.1 |
0.6 |
GO:0051533 |
positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.1 |
0.2 |
GO:0002447 |
eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) |
| 0.1 |
0.3 |
GO:0051639 |
actin filament network formation(GO:0051639) |
| 0.1 |
0.4 |
GO:0090209 |
negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.1 |
0.2 |
GO:0000733 |
DNA strand renaturation(GO:0000733) |
| 0.1 |
0.4 |
GO:0000395 |
mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 |
0.4 |
GO:1902416 |
positive regulation of mRNA binding(GO:1902416) positive regulation of RNA binding(GO:1905216) |
| 0.1 |
0.5 |
GO:0050774 |
negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.1 |
0.5 |
GO:0045899 |
positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 |
0.2 |
GO:0090365 |
regulation of mRNA modification(GO:0090365) |
| 0.1 |
0.7 |
GO:0072643 |
interferon-gamma secretion(GO:0072643) |
| 0.0 |
0.2 |
GO:0032026 |
response to magnesium ion(GO:0032026) |
| 0.0 |
0.5 |
GO:0097501 |
stress response to metal ion(GO:0097501) |
| 0.0 |
0.2 |
GO:0070236 |
positive regulation of immunoglobulin secretion(GO:0051024) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 |
0.7 |
GO:0008608 |
attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.0 |
0.5 |
GO:0032525 |
somite rostral/caudal axis specification(GO:0032525) |
| 0.0 |
0.1 |
GO:0042138 |
meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 |
0.4 |
GO:0042523 |
positive regulation of tyrosine phosphorylation of Stat5 protein(GO:0042523) |
| 0.0 |
0.5 |
GO:0007141 |
male meiosis I(GO:0007141) |
| 0.0 |
1.8 |
GO:0048013 |
ephrin receptor signaling pathway(GO:0048013) |
| 0.0 |
0.3 |
GO:0042769 |
DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 |
0.4 |
GO:0030238 |
male sex determination(GO:0030238) |
| 0.0 |
0.4 |
GO:0071257 |
cellular response to electrical stimulus(GO:0071257) |
| 0.0 |
0.2 |
GO:0031133 |
cellular magnesium ion homeostasis(GO:0010961) regulation of axon diameter(GO:0031133) |
| 0.0 |
1.3 |
GO:2001222 |
regulation of neuron migration(GO:2001222) |
| 0.0 |
0.4 |
GO:0021796 |
cerebral cortex regionalization(GO:0021796) |
| 0.0 |
0.3 |
GO:0033567 |
DNA replication, Okazaki fragment processing(GO:0033567) |
| 0.0 |
1.1 |
GO:1900087 |
positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 |
0.6 |
GO:0042407 |
cristae formation(GO:0042407) |
| 0.0 |
0.1 |
GO:0090234 |
regulation of centromere complex assembly(GO:0090230) regulation of kinetochore assembly(GO:0090234) |
| 0.0 |
0.2 |
GO:0006369 |
termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 |
0.1 |
GO:0019087 |
transformation of host cell by virus(GO:0019087) |
| 0.0 |
1.2 |
GO:0031103 |
axon regeneration(GO:0031103) |
| 0.0 |
0.6 |
GO:0048671 |
negative regulation of collateral sprouting(GO:0048671) |
| 0.0 |
0.7 |
GO:0033235 |
positive regulation of protein sumoylation(GO:0033235) |
| 0.0 |
1.0 |
GO:0015991 |
energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 |
0.3 |
GO:0071420 |
cellular response to histamine(GO:0071420) |
| 0.0 |
0.3 |
GO:2000392 |
regulation of lamellipodium morphogenesis(GO:2000392) positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.0 |
1.1 |
GO:0071277 |
cellular response to calcium ion(GO:0071277) |
| 0.0 |
0.5 |
GO:0015985 |
energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 |
0.1 |
GO:0070829 |
heterochromatin maintenance(GO:0070829) |
| 0.0 |
0.4 |
GO:0010388 |
protein deneddylation(GO:0000338) cullin deneddylation(GO:0010388) |
| 0.0 |
0.5 |
GO:0043968 |
histone H2A acetylation(GO:0043968) |
| 0.0 |
0.3 |
GO:0032486 |
Rap protein signal transduction(GO:0032486) |
| 0.0 |
0.6 |
GO:0008344 |
adult locomotory behavior(GO:0008344) |
| 0.0 |
0.3 |
GO:0010452 |
histone H3-K36 methylation(GO:0010452) |
| 0.0 |
0.7 |
GO:0030513 |
positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 |
0.3 |
GO:0043084 |
penile erection(GO:0043084) |
| 0.0 |
0.1 |
GO:0009169 |
purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 |
0.4 |
GO:0070584 |
mitochondrion morphogenesis(GO:0070584) |
| 0.0 |
0.6 |
GO:0034314 |
Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 |
0.4 |
GO:0006474 |
N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 |
0.3 |
GO:0034244 |
negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 |
0.4 |
GO:0050909 |
sensory perception of taste(GO:0050909) |
| 0.0 |
0.3 |
GO:0060670 |
branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 |
1.0 |
GO:0019228 |
neuronal action potential(GO:0019228) |
| 0.0 |
1.0 |
GO:0009142 |
nucleoside triphosphate biosynthetic process(GO:0009142) |
| 0.0 |
0.2 |
GO:0010667 |
negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 |
1.0 |
GO:0045669 |
positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 |
0.6 |
GO:0006270 |
DNA replication initiation(GO:0006270) |
| 0.0 |
0.2 |
GO:0009404 |
toxin metabolic process(GO:0009404) |
| 0.0 |
1.2 |
GO:0002088 |
lens development in camera-type eye(GO:0002088) |
| 0.0 |
0.4 |
GO:2001240 |
negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 |
0.4 |
GO:0006301 |
postreplication repair(GO:0006301) |
| 0.0 |
0.1 |
GO:0035743 |
CD4-positive, alpha-beta T cell cytokine production(GO:0035743) T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 |
3.0 |
GO:0043524 |
negative regulation of neuron apoptotic process(GO:0043524) |
| 0.0 |
0.4 |
GO:0006298 |
mismatch repair(GO:0006298) |
| 0.0 |
0.1 |
GO:0032536 |
regulation of cell projection size(GO:0032536) |
| 0.0 |
0.3 |
GO:0010614 |
negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 |
0.4 |
GO:0071480 |
cellular response to gamma radiation(GO:0071480) |
| 0.0 |
0.3 |
GO:0043044 |
ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 |
0.6 |
GO:0035690 |
cellular response to drug(GO:0035690) |
| 0.0 |
0.2 |
GO:0030150 |
protein import into mitochondrial matrix(GO:0030150) |
| 0.0 |
0.6 |
GO:0030970 |
retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 |
0.1 |
GO:0033600 |
negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 0.0 |
0.2 |
GO:0010225 |
response to UV-C(GO:0010225) |
| 0.0 |
0.3 |
GO:0043552 |
positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 |
0.7 |
GO:0006506 |
GPI anchor biosynthetic process(GO:0006506) |
| 0.0 |
0.3 |
GO:0007212 |
dopamine receptor signaling pathway(GO:0007212) |
| 0.0 |
0.8 |
GO:0055013 |
cardiac muscle cell development(GO:0055013) |
| 0.0 |
0.5 |
GO:0006890 |
retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 |
0.1 |
GO:0051189 |
Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 |
0.2 |
GO:0009408 |
response to heat(GO:0009408) |
| 0.0 |
0.1 |
GO:0048227 |
plasma membrane to endosome transport(GO:0048227) |
| 0.0 |
0.2 |
GO:0046597 |
negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 |
0.1 |
GO:1903361 |
protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 |
0.3 |
GO:0046677 |
response to antibiotic(GO:0046677) |
| 0.0 |
0.6 |
GO:0042073 |
intraciliary transport(GO:0042073) |
| 0.0 |
0.0 |
GO:2000383 |
regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.0 |
0.2 |
GO:2000816 |
negative regulation of mitotic metaphase/anaphase transition(GO:0045841) mitotic spindle checkpoint(GO:0071174) negative regulation of metaphase/anaphase transition of cell cycle(GO:1902100) negative regulation of mitotic sister chromatid separation(GO:2000816) |
| 0.0 |
0.1 |
GO:0045947 |
negative regulation of translational initiation(GO:0045947) |
| 0.0 |
0.4 |
GO:0045022 |
early endosome to late endosome transport(GO:0045022) |
| 0.0 |
0.3 |
GO:0070613 |
regulation of protein processing(GO:0070613) regulation of protein maturation(GO:1903317) |