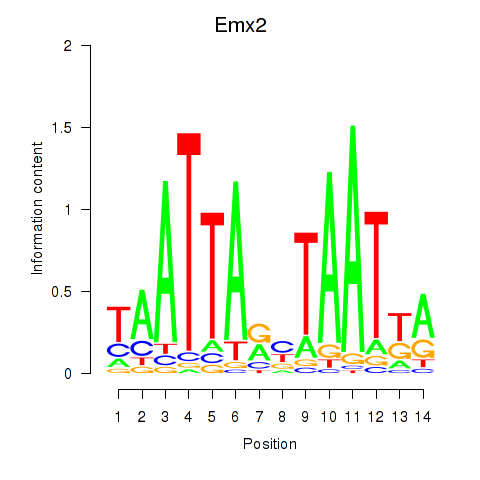
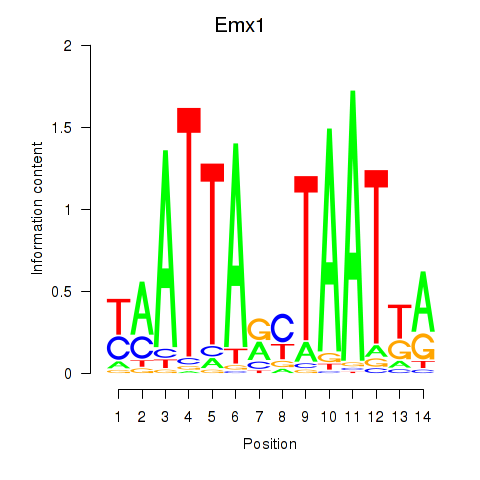
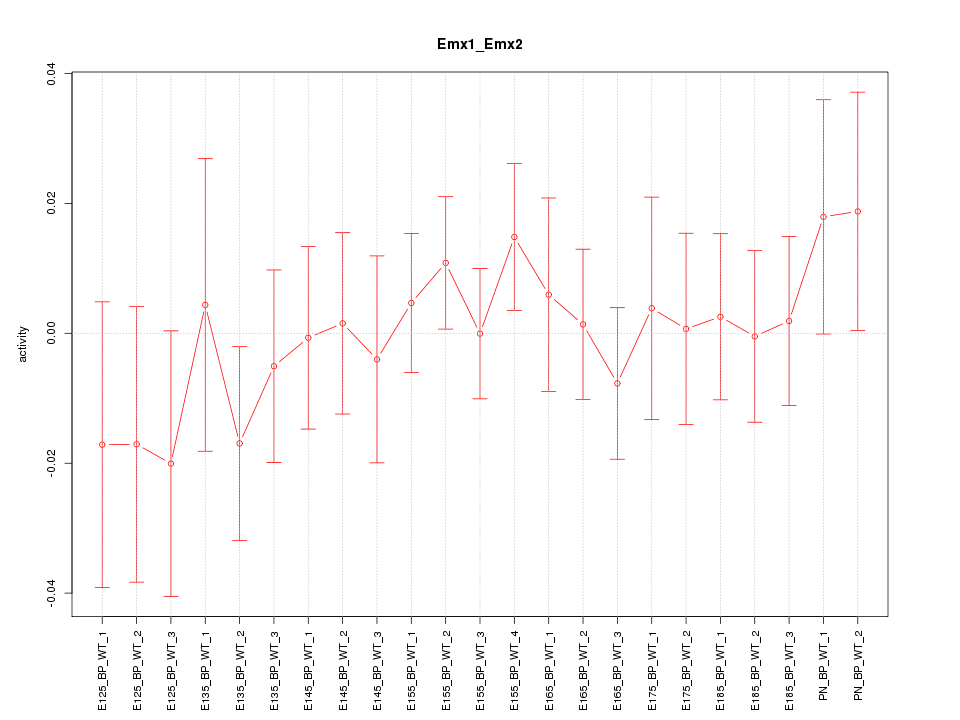
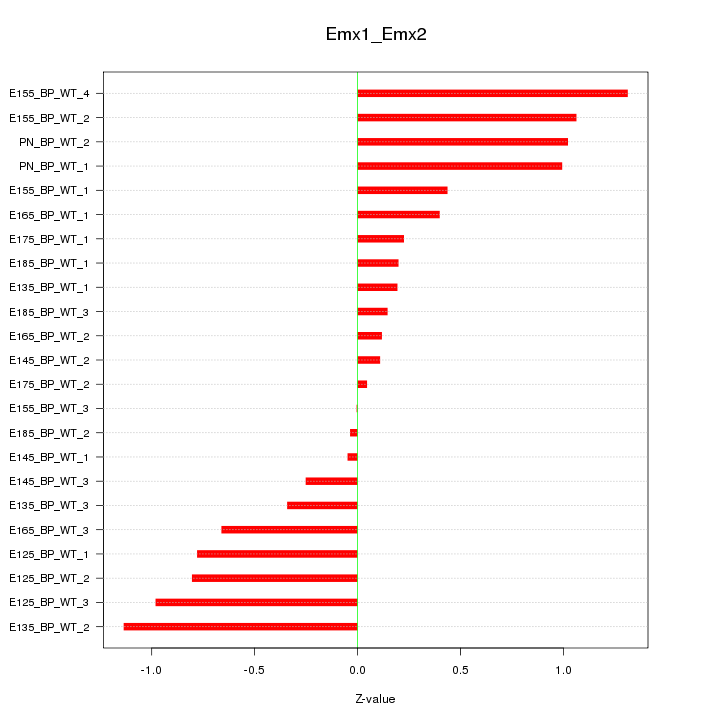
| 1.5 |
5.9 |
GO:2000795 |
negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.6 |
1.9 |
GO:2000564 |
CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) positive regulation of CD8-positive, alpha-beta T cell activation(GO:2001187) |
| 0.4 |
1.1 |
GO:0019343 |
cysteine biosynthetic process via cystathionine(GO:0019343) cysteine biosynthetic process(GO:0019344) |
| 0.3 |
1.9 |
GO:0016338 |
calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.2 |
0.8 |
GO:0014043 |
negative regulation of neuron maturation(GO:0014043) |
| 0.2 |
0.6 |
GO:0035633 |
maintenance of blood-brain barrier(GO:0035633) |
| 0.2 |
0.8 |
GO:0070295 |
renal water absorption(GO:0070295) |
| 0.1 |
0.8 |
GO:0060648 |
mammary gland bud morphogenesis(GO:0060648) |
| 0.1 |
1.1 |
GO:0038028 |
insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.1 |
0.5 |
GO:0035609 |
C-terminal protein deglutamylation(GO:0035609) |
| 0.1 |
0.7 |
GO:1902261 |
positive regulation of delayed rectifier potassium channel activity(GO:1902261) positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.1 |
1.2 |
GO:0030322 |
stabilization of membrane potential(GO:0030322) |
| 0.1 |
0.6 |
GO:0042636 |
negative regulation of hair cycle(GO:0042636) progesterone secretion(GO:0042701) negative regulation of hair follicle development(GO:0051799) positive regulation of ovulation(GO:0060279) |
| 0.1 |
0.3 |
GO:0071921 |
establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.1 |
1.2 |
GO:0015816 |
glycine transport(GO:0015816) |
| 0.1 |
0.5 |
GO:0042780 |
tRNA 3'-end processing(GO:0042780) |
| 0.1 |
1.1 |
GO:1900383 |
regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 |
0.6 |
GO:0015862 |
uridine transport(GO:0015862) |
| 0.1 |
0.3 |
GO:0050787 |
glycolate metabolic process(GO:0009441) enzyme active site formation via L-cysteine sulfinic acid(GO:0018323) primary alcohol biosynthetic process(GO:0034309) cellular response to glyoxal(GO:0036471) glycolate biosynthetic process(GO:0046295) detoxification of mercury ion(GO:0050787) negative regulation of TRAIL-activated apoptotic signaling pathway(GO:1903122) regulation of pyrroline-5-carboxylate reductase activity(GO:1903167) positive regulation of pyrroline-5-carboxylate reductase activity(GO:1903168) regulation of tyrosine 3-monooxygenase activity(GO:1903176) positive regulation of tyrosine 3-monooxygenase activity(GO:1903178) L-dopa metabolic process(GO:1903184) L-dopa biosynthetic process(GO:1903185) glyoxal metabolic process(GO:1903189) glyoxal catabolic process(GO:1903190) regulation of L-dopa biosynthetic process(GO:1903195) positive regulation of L-dopa biosynthetic process(GO:1903197) regulation of L-dopa decarboxylase activity(GO:1903198) positive regulation of L-dopa decarboxylase activity(GO:1903200) positive regulation of cellular amino acid biosynthetic process(GO:2000284) positive regulation of androgen receptor activity(GO:2000825) |
| 0.1 |
0.4 |
GO:0014719 |
skeletal muscle satellite cell activation(GO:0014719) |
| 0.1 |
0.2 |
GO:0060060 |
post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.1 |
1.1 |
GO:0016339 |
calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 |
0.6 |
GO:0033631 |
cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.1 |
0.2 |
GO:0051542 |
elastin biosynthetic process(GO:0051542) |
| 0.1 |
0.3 |
GO:0019236 |
response to pheromone(GO:0019236) |
| 0.0 |
0.1 |
GO:0006667 |
sphinganine metabolic process(GO:0006667) |
| 0.0 |
0.3 |
GO:0046477 |
glycosylceramide catabolic process(GO:0046477) |
| 0.0 |
0.4 |
GO:0036158 |
outer dynein arm assembly(GO:0036158) |
| 0.0 |
0.4 |
GO:0032020 |
ISG15-protein conjugation(GO:0032020) |
| 0.0 |
3.0 |
GO:0009063 |
cellular amino acid catabolic process(GO:0009063) |
| 0.0 |
0.3 |
GO:0006620 |
posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 |
2.0 |
GO:0038083 |
peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 |
0.4 |
GO:1901898 |
negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 |
0.3 |
GO:0045607 |
regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 |
0.1 |
GO:0021764 |
amygdala development(GO:0021764) |
| 0.0 |
0.2 |
GO:0045200 |
establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 |
0.1 |
GO:0031038 |
myosin II filament organization(GO:0031038) regulation of myosin II filament organization(GO:0043519) |
| 0.0 |
0.1 |
GO:0044028 |
DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.0 |
0.1 |
GO:0045113 |
regulation of integrin biosynthetic process(GO:0045113) |
| 0.0 |
0.6 |
GO:0042773 |
ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 |
0.1 |
GO:1901727 |
positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 |
0.1 |
GO:0032439 |
endosome localization(GO:0032439) |
| 0.0 |
0.1 |
GO:0034551 |
respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 |
0.2 |
GO:0033194 |
response to hydroperoxide(GO:0033194) |
| 0.0 |
0.1 |
GO:0045716 |
positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.0 |
0.5 |
GO:0007528 |
neuromuscular junction development(GO:0007528) |