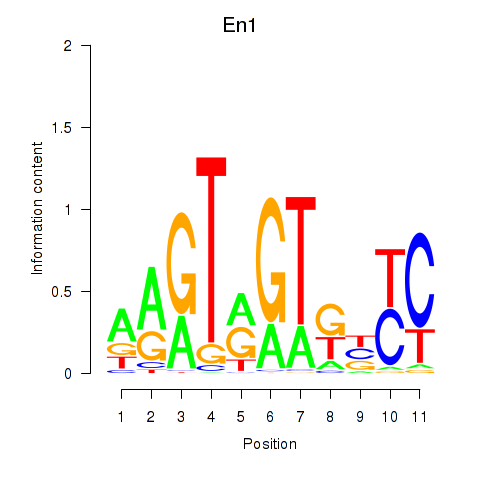
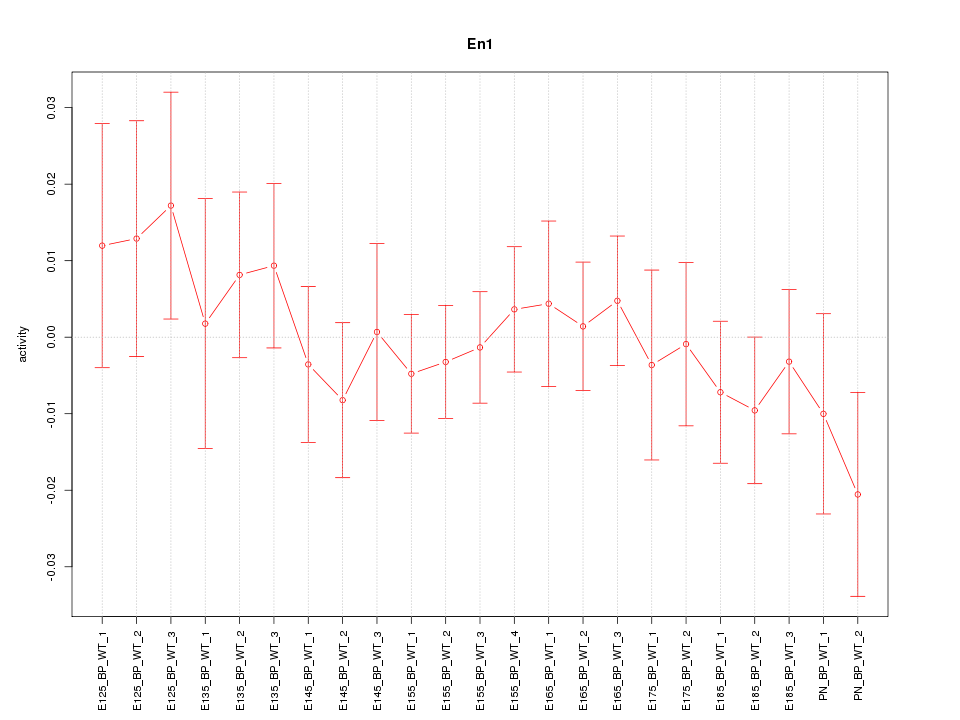
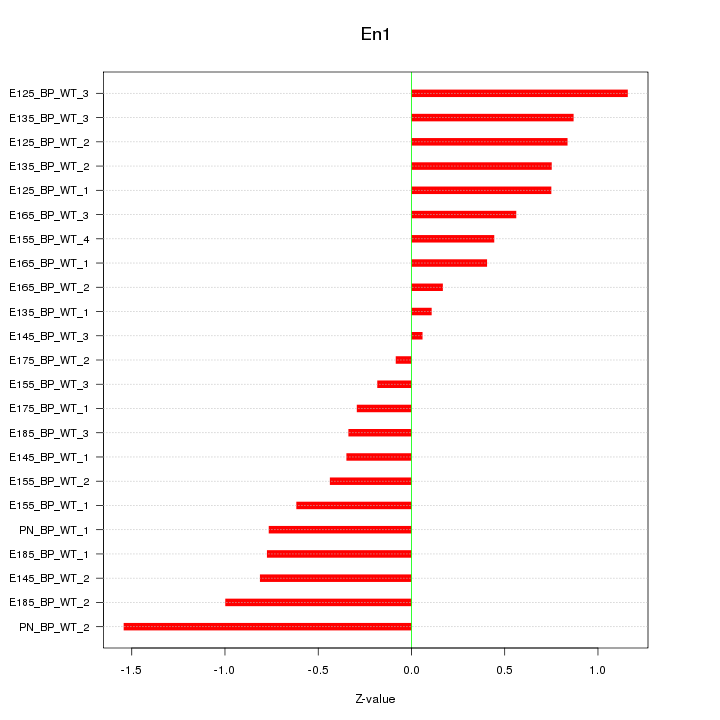
| 0.7 |
4.4 |
GO:0021797 |
forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.6 |
2.3 |
GO:1900623 |
positive regulation of keratinocyte proliferation(GO:0010838) regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.5 |
3.2 |
GO:1903690 |
negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.4 |
1.3 |
GO:0072070 |
establishment or maintenance of polarity of embryonic epithelium(GO:0016332) loop of Henle development(GO:0072070) metanephric loop of Henle development(GO:0072236) |
| 0.3 |
2.5 |
GO:0033564 |
anterior/posterior axon guidance(GO:0033564) |
| 0.3 |
0.9 |
GO:0042320 |
regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) circadian sleep/wake cycle, REM sleep(GO:0042747) positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 0.3 |
0.8 |
GO:0006436 |
tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.3 |
4.0 |
GO:0035563 |
positive regulation of chromatin binding(GO:0035563) |
| 0.3 |
0.8 |
GO:1904274 |
tricellular tight junction assembly(GO:1904274) |
| 0.2 |
1.5 |
GO:2000210 |
positive regulation of anoikis(GO:2000210) |
| 0.2 |
0.2 |
GO:0072425 |
signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.2 |
0.8 |
GO:0045091 |
regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.2 |
0.9 |
GO:0033087 |
negative regulation of immature T cell proliferation(GO:0033087) |
| 0.2 |
0.5 |
GO:0002636 |
positive regulation of germinal center formation(GO:0002636) |
| 0.2 |
0.2 |
GO:0039692 |
single stranded viral RNA replication via double stranded DNA intermediate(GO:0039692) |
| 0.2 |
2.4 |
GO:0032211 |
negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.2 |
0.5 |
GO:0048162 |
preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.1 |
0.4 |
GO:0097278 |
complement-dependent cytotoxicity(GO:0097278) |
| 0.1 |
0.3 |
GO:0072048 |
pattern specification involved in kidney development(GO:0061004) renal system pattern specification(GO:0072048) |
| 0.1 |
0.4 |
GO:1903896 |
positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.1 |
2.0 |
GO:0060052 |
neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 |
0.6 |
GO:0070829 |
heterochromatin maintenance(GO:0070829) |
| 0.1 |
0.3 |
GO:0070487 |
monocyte aggregation(GO:0070487) |
| 0.1 |
0.3 |
GO:0072554 |
blood vessel lumenization(GO:0072554) |
| 0.1 |
1.1 |
GO:0033353 |
S-adenosylmethionine cycle(GO:0033353) |
| 0.1 |
0.4 |
GO:0001951 |
intestinal D-glucose absorption(GO:0001951) terminal web assembly(GO:1902896) |
| 0.1 |
0.7 |
GO:0060467 |
negative regulation of fertilization(GO:0060467) |
| 0.1 |
0.7 |
GO:0032229 |
negative regulation of synaptic transmission, GABAergic(GO:0032229) |
| 0.1 |
0.3 |
GO:0002014 |
vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.1 |
0.9 |
GO:0035542 |
regulation of SNARE complex assembly(GO:0035542) |
| 0.1 |
0.4 |
GO:0002322 |
B cell proliferation involved in immune response(GO:0002322) |
| 0.1 |
0.5 |
GO:0051852 |
antifungal humoral response(GO:0019732) disruption by host of symbiont cells(GO:0051852) killing by host of symbiont cells(GO:0051873) |
| 0.1 |
0.4 |
GO:0035616 |
histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.1 |
0.8 |
GO:0070474 |
positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.1 |
0.4 |
GO:0043490 |
malate-aspartate shuttle(GO:0043490) |
| 0.1 |
0.5 |
GO:0030091 |
protein repair(GO:0030091) |
| 0.1 |
1.8 |
GO:0043567 |
regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 |
0.4 |
GO:1904357 |
negative regulation of telomere maintenance via telomere lengthening(GO:1904357) |
| 0.1 |
3.5 |
GO:0045746 |
negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 |
0.3 |
GO:0019659 |
glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.1 |
0.3 |
GO:1902277 |
negative regulation of pancreatic amylase secretion(GO:1902277) |
| 0.1 |
2.9 |
GO:0006270 |
DNA replication initiation(GO:0006270) |
| 0.1 |
0.3 |
GO:1901524 |
regulation of macromitophagy(GO:1901524) negative regulation of macromitophagy(GO:1901525) |
| 0.1 |
0.3 |
GO:0070358 |
actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 |
1.9 |
GO:0008340 |
determination of adult lifespan(GO:0008340) |
| 0.1 |
0.5 |
GO:0006957 |
complement activation, alternative pathway(GO:0006957) |
| 0.1 |
0.2 |
GO:0010986 |
complement receptor mediated signaling pathway(GO:0002430) GPI anchor release(GO:0006507) positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.1 |
0.7 |
GO:0046085 |
adenosine metabolic process(GO:0046085) |
| 0.1 |
0.4 |
GO:0046600 |
negative regulation of centriole replication(GO:0046600) |
| 0.1 |
0.5 |
GO:1900194 |
negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.1 |
0.3 |
GO:0070417 |
cellular response to cold(GO:0070417) |
| 0.1 |
0.5 |
GO:0044351 |
macropinocytosis(GO:0044351) |
| 0.1 |
1.0 |
GO:0016188 |
synaptic vesicle maturation(GO:0016188) |
| 0.1 |
0.4 |
GO:0071883 |
activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.1 |
0.5 |
GO:0006977 |
DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.1 |
0.2 |
GO:0072592 |
oxygen metabolic process(GO:0072592) |
| 0.1 |
0.4 |
GO:0046602 |
regulation of mitotic centrosome separation(GO:0046602) |
| 0.1 |
0.2 |
GO:1900017 |
positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.1 |
0.4 |
GO:0018243 |
protein O-linked glycosylation via threonine(GO:0018243) |
| 0.1 |
0.2 |
GO:0010920 |
negative regulation of inositol phosphate biosynthetic process(GO:0010920) |
| 0.1 |
0.5 |
GO:0035871 |
protein K11-linked deubiquitination(GO:0035871) |
| 0.1 |
0.5 |
GO:0050832 |
defense response to fungus(GO:0050832) |
| 0.0 |
0.2 |
GO:0031581 |
hemidesmosome assembly(GO:0031581) |
| 0.0 |
0.1 |
GO:0010636 |
positive regulation of mitochondrial fusion(GO:0010636) |
| 0.0 |
0.3 |
GO:0006995 |
cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 |
0.3 |
GO:0001842 |
neural fold formation(GO:0001842) |
| 0.0 |
0.5 |
GO:2001256 |
regulation of store-operated calcium entry(GO:2001256) |
| 0.0 |
0.4 |
GO:0043353 |
enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 |
0.4 |
GO:0051967 |
negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 |
0.7 |
GO:0048387 |
negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 |
0.6 |
GO:0050849 |
negative regulation of calcium-mediated signaling(GO:0050849) |
| 0.0 |
0.1 |
GO:0048296 |
isotype switching to IgA isotypes(GO:0048290) regulation of isotype switching to IgA isotypes(GO:0048296) |
| 0.0 |
0.3 |
GO:0031339 |
negative regulation of vesicle fusion(GO:0031339) |
| 0.0 |
0.8 |
GO:0007263 |
nitric oxide mediated signal transduction(GO:0007263) |
| 0.0 |
0.2 |
GO:1904263 |
positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 |
0.2 |
GO:0042985 |
negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 |
0.3 |
GO:0019348 |
dolichol-linked oligosaccharide biosynthetic process(GO:0006488) dolichol metabolic process(GO:0019348) |
| 0.0 |
0.4 |
GO:0007175 |
negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 |
0.4 |
GO:2000643 |
positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.0 |
0.3 |
GO:0090043 |
regulation of tubulin deacetylation(GO:0090043) |
| 0.0 |
0.5 |
GO:0007519 |
skeletal muscle tissue development(GO:0007519) |
| 0.0 |
0.5 |
GO:0030574 |
collagen catabolic process(GO:0030574) negative regulation of phagocytosis(GO:0050765) |
| 0.0 |
0.1 |
GO:1900454 |
positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 |
0.2 |
GO:1904491 |
protein localization to ciliary transition zone(GO:1904491) |
| 0.0 |
0.6 |
GO:0008209 |
androgen metabolic process(GO:0008209) |
| 0.0 |
0.1 |
GO:0036092 |
phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 |
0.1 |
GO:0007171 |
activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) prolactin signaling pathway(GO:0038161) |
| 0.0 |
0.1 |
GO:1901723 |
negative regulation of cell proliferation involved in kidney development(GO:1901723) negative regulation of cellular respiration(GO:1901856) |
| 0.0 |
1.4 |
GO:0045773 |
positive regulation of axon extension(GO:0045773) |
| 0.0 |
0.6 |
GO:0030308 |
negative regulation of cell growth(GO:0030308) |
| 0.0 |
1.7 |
GO:0030512 |
negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 |
0.2 |
GO:0010747 |
positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.0 |
1.0 |
GO:1902476 |
chloride transmembrane transport(GO:1902476) |
| 0.0 |
0.3 |
GO:0051764 |
actin crosslink formation(GO:0051764) |
| 0.0 |
0.2 |
GO:0033327 |
Leydig cell differentiation(GO:0033327) |
| 0.0 |
0.5 |
GO:0014829 |
vascular smooth muscle contraction(GO:0014829) |
| 0.0 |
0.0 |
GO:1904468 |
negative regulation of interleukin-1 beta secretion(GO:0050713) negative regulation of tumor necrosis factor secretion(GO:1904468) |
| 0.0 |
0.1 |
GO:0031119 |
tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 |
0.1 |
GO:0048023 |
positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 |
0.0 |
GO:0032792 |
negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.0 |
0.3 |
GO:0031571 |
mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.0 |
0.1 |
GO:0016191 |
synaptic vesicle uncoating(GO:0016191) regulation of endosome organization(GO:1904978) positive regulation of endosome organization(GO:1904980) |
| 0.0 |
0.4 |
GO:0032981 |
NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 |
0.4 |
GO:0016339 |
calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 |
0.1 |
GO:0051225 |
spindle assembly(GO:0051225) |
| 0.0 |
0.2 |
GO:1902018 |
negative regulation of cilium assembly(GO:1902018) |
| 0.0 |
0.2 |
GO:0007221 |
positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 |
0.1 |
GO:0060011 |
Sertoli cell proliferation(GO:0060011) |
| 0.0 |
0.3 |
GO:0006825 |
copper ion transport(GO:0006825) |
| 0.0 |
0.5 |
GO:0097120 |
receptor localization to synapse(GO:0097120) |
| 0.0 |
0.2 |
GO:0006614 |
SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 |
0.4 |
GO:0060348 |
bone development(GO:0060348) |
| 0.0 |
0.1 |
GO:0019372 |
lipoxygenase pathway(GO:0019372) |
| 0.0 |
0.4 |
GO:0001782 |
B cell homeostasis(GO:0001782) |
| 0.0 |
0.0 |
GO:0035521 |
monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 |
0.3 |
GO:0006828 |
manganese ion transport(GO:0006828) |
| 0.0 |
0.0 |
GO:0006553 |
lysine metabolic process(GO:0006553) |
| 0.0 |
1.3 |
GO:0060349 |
bone morphogenesis(GO:0060349) |
| 0.0 |
0.2 |
GO:0035020 |
regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 |
0.4 |
GO:0008608 |
attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.0 |
0.1 |
GO:0046036 |
GTP biosynthetic process(GO:0006183) CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.0 |
0.9 |
GO:0007156 |
homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 |
0.1 |
GO:1903975 |
regulation of glial cell migration(GO:1903975) |
| 0.0 |
0.7 |
GO:0030901 |
midbrain development(GO:0030901) |
| 0.0 |
0.2 |
GO:0036065 |
fucosylation(GO:0036065) |
| 0.0 |
0.4 |
GO:0000470 |
maturation of LSU-rRNA(GO:0000470) |
| 0.0 |
0.2 |
GO:0001504 |
neurotransmitter uptake(GO:0001504) |
| 0.0 |
0.2 |
GO:1901741 |
positive regulation of myoblast fusion(GO:1901741) |
| 0.0 |
0.3 |
GO:0033622 |
integrin activation(GO:0033622) |
| 0.0 |
0.3 |
GO:0045724 |
positive regulation of cilium assembly(GO:0045724) |
| 0.0 |
0.0 |
GO:0046826 |
negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 |
0.5 |
GO:0045668 |
negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 |
0.1 |
GO:0010758 |
regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 |
0.5 |
GO:0006376 |
mRNA splice site selection(GO:0006376) |
| 0.0 |
0.7 |
GO:0030968 |
endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 |
0.0 |
GO:0032877 |
positive regulation of DNA endoreduplication(GO:0032877) |
| 0.0 |
0.1 |
GO:0035093 |
spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 |
0.7 |
GO:0016574 |
histone ubiquitination(GO:0016574) |
| 0.0 |
0.1 |
GO:0090503 |
RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 |
0.0 |
GO:0050915 |
sensory perception of sour taste(GO:0050915) |
| 0.0 |
0.3 |
GO:0048488 |
synaptic vesicle endocytosis(GO:0048488) |
| 0.0 |
0.7 |
GO:0046847 |
filopodium assembly(GO:0046847) |
| 0.0 |
1.0 |
GO:0001843 |
neural tube closure(GO:0001843) primary neural tube formation(GO:0014020) tube closure(GO:0060606) |