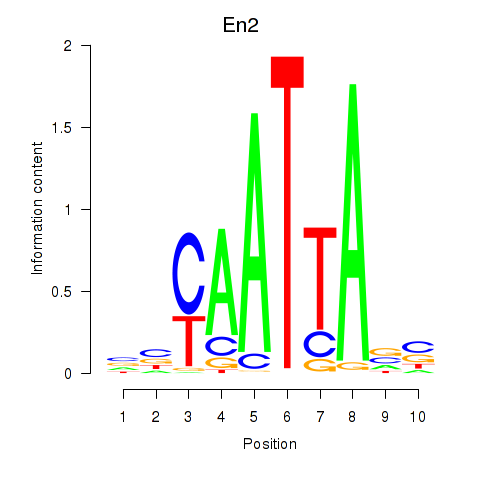
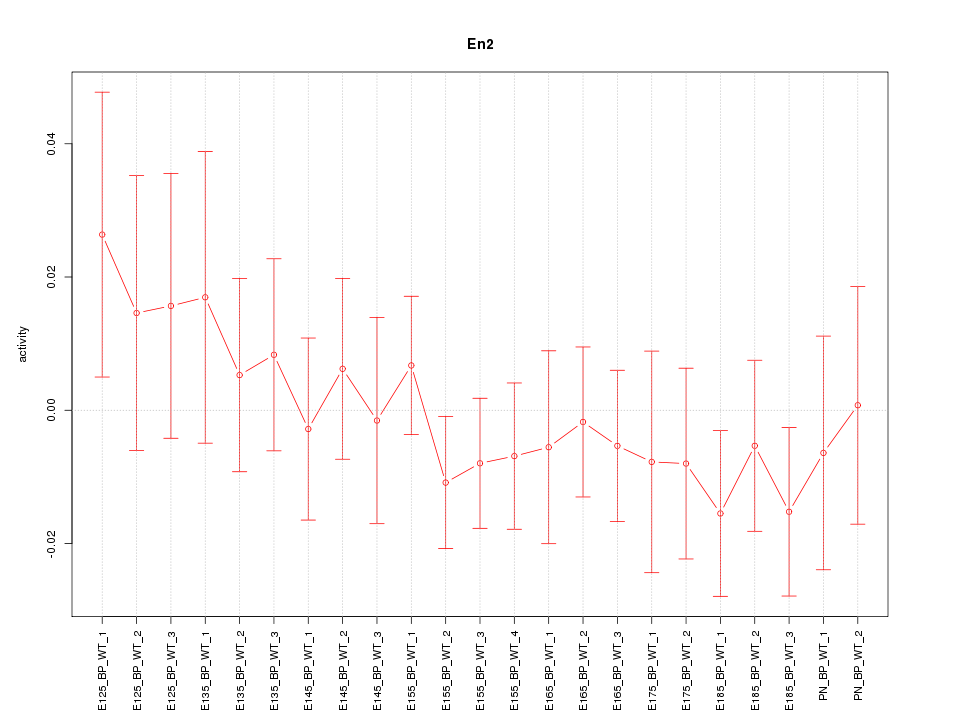
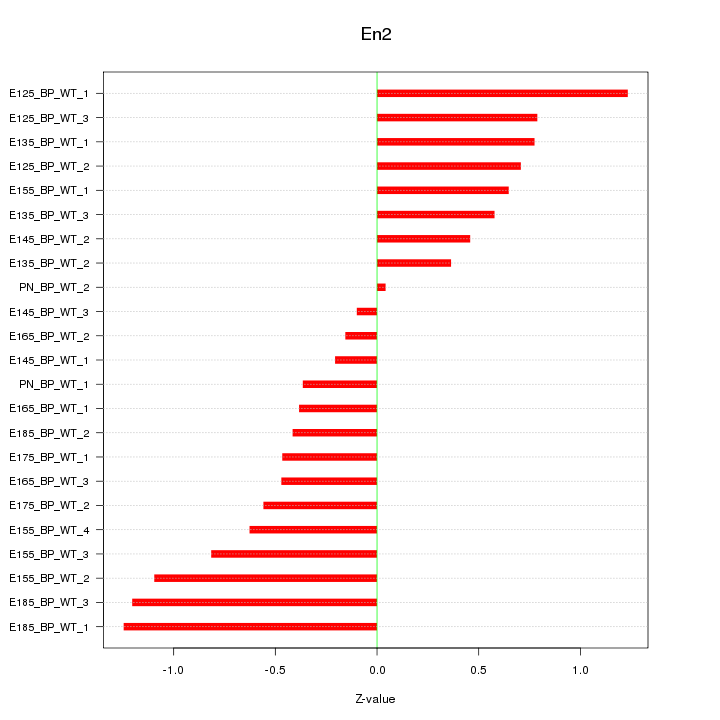
| 1.3 |
3.9 |
GO:0090274 |
positive regulation of somatostatin secretion(GO:0090274) |
| 0.7 |
5.9 |
GO:0035726 |
common myeloid progenitor cell proliferation(GO:0035726) |
| 0.5 |
3.3 |
GO:0051798 |
positive regulation of hair follicle development(GO:0051798) |
| 0.4 |
1.1 |
GO:0060023 |
soft palate development(GO:0060023) |
| 0.3 |
1.4 |
GO:0051866 |
general adaptation syndrome(GO:0051866) |
| 0.3 |
0.9 |
GO:2000393 |
negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.2 |
3.9 |
GO:0030953 |
astral microtubule organization(GO:0030953) |
| 0.2 |
0.7 |
GO:0030327 |
prenylated protein catabolic process(GO:0030327) |
| 0.2 |
0.7 |
GO:0042536 |
negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.2 |
1.0 |
GO:0048245 |
eosinophil chemotaxis(GO:0048245) |
| 0.2 |
0.6 |
GO:0002669 |
positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.2 |
0.7 |
GO:0061055 |
myotome development(GO:0061055) |
| 0.2 |
0.5 |
GO:0036091 |
positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.2 |
0.7 |
GO:2001032 |
regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.2 |
0.5 |
GO:0007290 |
spermatid nucleus elongation(GO:0007290) |
| 0.2 |
3.0 |
GO:0007530 |
sex determination(GO:0007530) |
| 0.2 |
1.6 |
GO:0016198 |
axon choice point recognition(GO:0016198) |
| 0.2 |
0.5 |
GO:0045113 |
regulation of integrin biosynthetic process(GO:0045113) |
| 0.2 |
2.8 |
GO:0051988 |
regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.2 |
1.2 |
GO:0005513 |
detection of calcium ion(GO:0005513) |
| 0.1 |
1.3 |
GO:0050957 |
equilibrioception(GO:0050957) |
| 0.1 |
0.6 |
GO:0032512 |
regulation of protein phosphatase type 2B activity(GO:0032512) positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.1 |
2.2 |
GO:0000920 |
cell separation after cytokinesis(GO:0000920) |
| 0.1 |
1.0 |
GO:0046645 |
positive regulation of gamma-delta T cell differentiation(GO:0045588) positive regulation of gamma-delta T cell activation(GO:0046645) positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.1 |
0.5 |
GO:1900028 |
negative regulation of ruffle assembly(GO:1900028) |
| 0.1 |
3.5 |
GO:0034723 |
DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 |
0.3 |
GO:1904879 |
positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.1 |
0.4 |
GO:0001550 |
ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 |
1.5 |
GO:0021796 |
cerebral cortex regionalization(GO:0021796) |
| 0.1 |
1.1 |
GO:0019985 |
translesion synthesis(GO:0019985) |
| 0.1 |
0.7 |
GO:0031547 |
brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.1 |
1.9 |
GO:0008340 |
determination of adult lifespan(GO:0008340) |
| 0.1 |
0.2 |
GO:0030300 |
regulation of intestinal cholesterol absorption(GO:0030300) |
| 0.1 |
2.6 |
GO:0007638 |
mechanosensory behavior(GO:0007638) |
| 0.1 |
0.5 |
GO:0007021 |
tubulin complex assembly(GO:0007021) |
| 0.1 |
0.8 |
GO:0031017 |
exocrine pancreas development(GO:0031017) |
| 0.1 |
1.0 |
GO:0099625 |
regulation of ventricular cardiac muscle cell membrane repolarization(GO:0060307) ventricular cardiac muscle cell membrane repolarization(GO:0099625) |
| 0.1 |
0.2 |
GO:0071609 |
negative regulation of interleukin-17 production(GO:0032700) neutrophil mediated killing of bacterium(GO:0070944) chemokine (C-C motif) ligand 5 production(GO:0071609) |
| 0.0 |
0.3 |
GO:0034638 |
phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 |
0.5 |
GO:1903504 |
regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 |
0.3 |
GO:0043415 |
positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.0 |
0.8 |
GO:0045672 |
positive regulation of osteoclast differentiation(GO:0045672) |
| 0.0 |
0.1 |
GO:0002578 |
negative regulation of antigen processing and presentation(GO:0002578) |
| 0.0 |
1.7 |
GO:0035307 |
positive regulation of protein dephosphorylation(GO:0035307) |
| 0.0 |
0.9 |
GO:0009409 |
response to cold(GO:0009409) |
| 0.0 |
0.5 |
GO:2000114 |
regulation of establishment of cell polarity(GO:2000114) |
| 0.0 |
0.3 |
GO:0001973 |
adenosine receptor signaling pathway(GO:0001973) |
| 0.0 |
0.2 |
GO:0045218 |
zonula adherens maintenance(GO:0045218) |
| 0.0 |
0.1 |
GO:0010847 |
regulation of chromatin assembly(GO:0010847) |
| 0.0 |
0.2 |
GO:0070562 |
regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 |
0.3 |
GO:0006120 |
mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 |
0.7 |
GO:1902236 |
negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.0 |
0.7 |
GO:0034508 |
centromere complex assembly(GO:0034508) |
| 0.0 |
0.4 |
GO:0043968 |
histone H2A acetylation(GO:0043968) |
| 0.0 |
0.6 |
GO:0015985 |
energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 |
1.5 |
GO:0030512 |
negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 |
0.3 |
GO:0006107 |
oxaloacetate metabolic process(GO:0006107) |
| 0.0 |
1.7 |
GO:0050885 |
neuromuscular process controlling balance(GO:0050885) |
| 0.0 |
0.1 |
GO:0097167 |
circadian regulation of translation(GO:0097167) |
| 0.0 |
0.3 |
GO:0051894 |
positive regulation of focal adhesion assembly(GO:0051894) |
| 0.0 |
0.2 |
GO:0032486 |
Rap protein signal transduction(GO:0032486) |
| 0.0 |
0.4 |
GO:0000266 |
mitochondrial fission(GO:0000266) |
| 0.0 |
0.7 |
GO:1904893 |
negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 |
0.8 |
GO:0006334 |
nucleosome assembly(GO:0006334) |
| 0.0 |
0.3 |
GO:0071384 |
cellular response to corticosteroid stimulus(GO:0071384) cellular response to glucocorticoid stimulus(GO:0071385) |