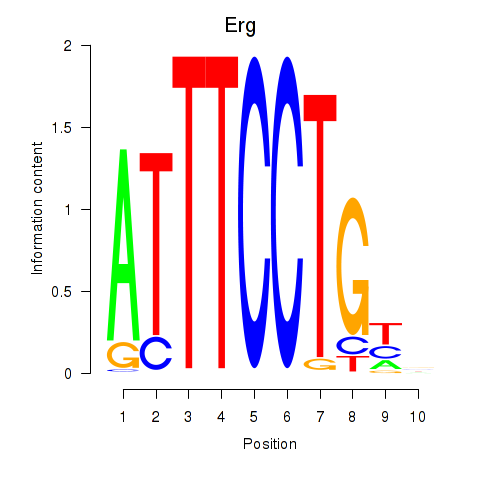
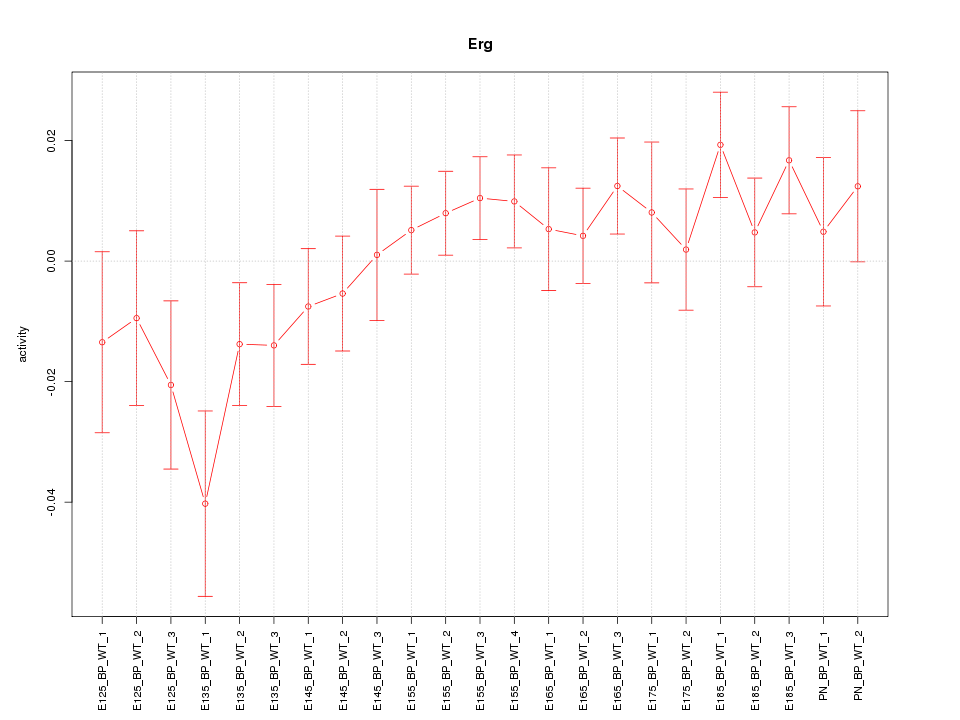
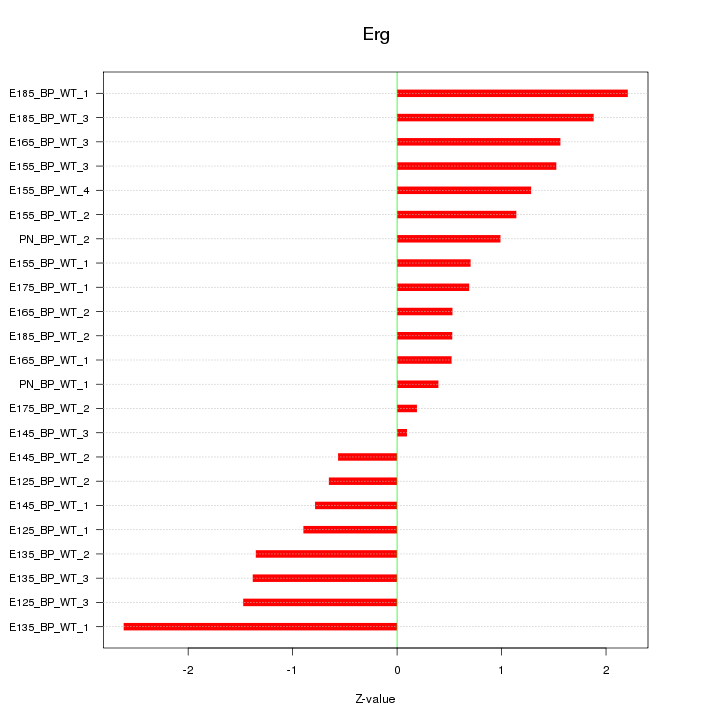
| 1.8 |
20.3 |
GO:0042572 |
retinol metabolic process(GO:0042572) |
| 1.2 |
1.2 |
GO:0038091 |
VEGF-activated platelet-derived growth factor receptor signaling pathway(GO:0038086) positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway(GO:0038091) |
| 1.0 |
2.9 |
GO:0003032 |
detection of oxygen(GO:0003032) |
| 0.8 |
0.8 |
GO:0043313 |
regulation of neutrophil degranulation(GO:0043313) regulation of neutrophil activation(GO:1902563) |
| 0.8 |
12.2 |
GO:0021902 |
commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.7 |
2.2 |
GO:0035441 |
cell migration involved in vasculogenesis(GO:0035441) neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) positive regulation of retinal ganglion cell axon guidance(GO:1902336) |
| 0.7 |
3.5 |
GO:0072015 |
glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.7 |
2.1 |
GO:0042939 |
renal sodium ion transport(GO:0003096) glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.7 |
2.0 |
GO:1900020 |
Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) hypophysis morphogenesis(GO:0048850) cervix development(GO:0060067) planar cell polarity pathway involved in outflow tract morphogenesis(GO:0061347) planar cell polarity pathway involved in ventricular septum morphogenesis(GO:0061348) planar cell polarity pathway involved in cardiac right atrium morphogenesis(GO:0061349) planar cell polarity pathway involved in cardiac muscle tissue morphogenesis(GO:0061350) planar cell polarity pathway involved in pericardium morphogenesis(GO:0061354) regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.6 |
2.8 |
GO:0032497 |
detection of lipopolysaccharide(GO:0032497) |
| 0.5 |
0.5 |
GO:0038163 |
thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.5 |
2.2 |
GO:0050904 |
diapedesis(GO:0050904) |
| 0.5 |
2.3 |
GO:0003199 |
endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.5 |
1.8 |
GO:0051562 |
negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.4 |
2.1 |
GO:0036438 |
maintenance of lens transparency(GO:0036438) |
| 0.4 |
2.1 |
GO:0060762 |
regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.4 |
3.7 |
GO:0061299 |
retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.4 |
1.1 |
GO:0035604 |
fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) coronal suture morphogenesis(GO:0060365) squamous basal epithelial stem cell differentiation involved in prostate gland acinus development(GO:0060529) |
| 0.4 |
0.4 |
GO:0030947 |
regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030947) |
| 0.3 |
1.7 |
GO:0006447 |
regulation of translational initiation by iron(GO:0006447) positive regulation of translational initiation by iron(GO:0045994) |
| 0.3 |
2.0 |
GO:0002281 |
macrophage activation involved in immune response(GO:0002281) |
| 0.3 |
2.9 |
GO:0090050 |
positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.3 |
0.9 |
GO:0071544 |
diphosphoinositol polyphosphate metabolic process(GO:0071543) diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.3 |
0.9 |
GO:0060913 |
stem cell fate commitment(GO:0048865) stem cell fate specification(GO:0048866) cardiac cell fate determination(GO:0060913) regulation of cardiac cell fate specification(GO:2000043) |
| 0.3 |
1.1 |
GO:1904425 |
negative regulation of GTP binding(GO:1904425) |
| 0.3 |
0.8 |
GO:0070944 |
neutrophil mediated killing of bacterium(GO:0070944) |
| 0.3 |
9.2 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.3 |
1.3 |
GO:0010701 |
positive regulation of norepinephrine secretion(GO:0010701) positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.3 |
1.3 |
GO:2000574 |
regulation of microtubule motor activity(GO:2000574) |
| 0.2 |
1.0 |
GO:1990743 |
protein sialylation(GO:1990743) |
| 0.2 |
1.7 |
GO:0001955 |
blood vessel maturation(GO:0001955) |
| 0.2 |
0.7 |
GO:0007161 |
calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.2 |
1.4 |
GO:1901534 |
post-embryonic camera-type eye morphogenesis(GO:0048597) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.2 |
3.6 |
GO:0051150 |
regulation of smooth muscle cell differentiation(GO:0051150) |
| 0.2 |
2.8 |
GO:1901898 |
negative regulation of relaxation of muscle(GO:1901078) regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.2 |
0.9 |
GO:0019276 |
UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.2 |
3.0 |
GO:1902287 |
semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.2 |
0.4 |
GO:0043323 |
positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.2 |
1.6 |
GO:0007016 |
cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.2 |
1.4 |
GO:0023035 |
CD40 signaling pathway(GO:0023035) |
| 0.2 |
0.6 |
GO:0035526 |
retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.2 |
0.8 |
GO:0016081 |
synaptic vesicle docking(GO:0016081) |
| 0.2 |
2.1 |
GO:1901621 |
negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.2 |
1.1 |
GO:1903352 |
ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.2 |
0.5 |
GO:0031086 |
nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.2 |
0.7 |
GO:2000211 |
regulation of glutamate metabolic process(GO:2000211) |
| 0.2 |
1.6 |
GO:0060613 |
fat pad development(GO:0060613) |
| 0.2 |
0.6 |
GO:1903237 |
negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.2 |
0.5 |
GO:0035435 |
phosphate ion transmembrane transport(GO:0035435) |
| 0.2 |
0.6 |
GO:0043654 |
recognition of apoptotic cell(GO:0043654) |
| 0.1 |
0.6 |
GO:0006867 |
asparagine transport(GO:0006867) glutamine transport(GO:0006868) |
| 0.1 |
0.7 |
GO:0006680 |
glucosylceramide catabolic process(GO:0006680) |
| 0.1 |
1.0 |
GO:0048102 |
autophagic cell death(GO:0048102) |
| 0.1 |
0.8 |
GO:0002681 |
somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.1 |
0.4 |
GO:0032241 |
positive regulation of nucleobase-containing compound transport(GO:0032241) regulation of nucleoside transport(GO:0032242) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 0.1 |
0.8 |
GO:0016557 |
peroxisome membrane biogenesis(GO:0016557) regulation of peroxisome size(GO:0044375) |
| 0.1 |
1.8 |
GO:0034497 |
protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 |
0.8 |
GO:0046880 |
regulation of follicle-stimulating hormone secretion(GO:0046880) follicle-stimulating hormone secretion(GO:0046884) |
| 0.1 |
0.4 |
GO:0033364 |
mast cell secretory granule organization(GO:0033364) |
| 0.1 |
0.5 |
GO:0042364 |
water-soluble vitamin biosynthetic process(GO:0042364) positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.1 |
1.2 |
GO:2001054 |
negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.1 |
0.5 |
GO:1901668 |
regulation of superoxide dismutase activity(GO:1901668) |
| 0.1 |
0.5 |
GO:0010726 |
positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.1 |
0.6 |
GO:1902774 |
late endosome to lysosome transport(GO:1902774) |
| 0.1 |
0.4 |
GO:0006776 |
vitamin A metabolic process(GO:0006776) |
| 0.1 |
2.1 |
GO:0030213 |
hyaluronan biosynthetic process(GO:0030213) |
| 0.1 |
0.6 |
GO:0014022 |
neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.1 |
1.8 |
GO:0043011 |
myeloid dendritic cell differentiation(GO:0043011) |
| 0.1 |
0.5 |
GO:0060842 |
arterial endothelial cell differentiation(GO:0060842) |
| 0.1 |
3.0 |
GO:0045026 |
plasma membrane fusion(GO:0045026) |
| 0.1 |
0.3 |
GO:0098501 |
polynucleotide dephosphorylation(GO:0098501) |
| 0.1 |
0.5 |
GO:0007089 |
traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.1 |
0.3 |
GO:0006624 |
vacuolar protein processing(GO:0006624) |
| 0.1 |
0.5 |
GO:0002051 |
osteoblast fate commitment(GO:0002051) |
| 0.1 |
0.7 |
GO:0042511 |
positive regulation of tyrosine phosphorylation of Stat1 protein(GO:0042511) |
| 0.1 |
0.4 |
GO:1903966 |
monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.1 |
0.3 |
GO:0048294 |
negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.1 |
0.3 |
GO:2000832 |
negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.1 |
1.4 |
GO:0070932 |
histone H3 deacetylation(GO:0070932) |
| 0.1 |
0.2 |
GO:0061010 |
gall bladder development(GO:0061010) |
| 0.1 |
1.0 |
GO:2000671 |
regulation of motor neuron apoptotic process(GO:2000671) |
| 0.1 |
0.2 |
GO:0070425 |
negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) |
| 0.1 |
1.0 |
GO:0070574 |
cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.1 |
1.6 |
GO:0050765 |
negative regulation of phagocytosis(GO:0050765) |
| 0.1 |
0.2 |
GO:0033278 |
cell proliferation in midbrain(GO:0033278) |
| 0.1 |
0.7 |
GO:0036462 |
TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.1 |
1.0 |
GO:0071225 |
cellular response to muramyl dipeptide(GO:0071225) |
| 0.1 |
1.0 |
GO:0010447 |
response to acidic pH(GO:0010447) |
| 0.1 |
0.7 |
GO:0006013 |
mannose metabolic process(GO:0006013) |
| 0.1 |
0.4 |
GO:1900748 |
positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 |
0.4 |
GO:0048149 |
behavioral response to ethanol(GO:0048149) |
| 0.1 |
0.4 |
GO:0021993 |
initiation of neural tube closure(GO:0021993) |
| 0.1 |
0.7 |
GO:0061088 |
sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 |
0.4 |
GO:0038032 |
termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.1 |
0.6 |
GO:0000022 |
mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.1 |
0.4 |
GO:0090273 |
regulation of somatostatin secretion(GO:0090273) |
| 0.1 |
2.8 |
GO:0001937 |
negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.1 |
4.5 |
GO:2001238 |
positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.1 |
0.2 |
GO:0034447 |
very-low-density lipoprotein particle clearance(GO:0034447) |
| 0.1 |
0.3 |
GO:0090309 |
positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.1 |
1.6 |
GO:0034340 |
response to type I interferon(GO:0034340) |
| 0.1 |
0.3 |
GO:0030397 |
membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.1 |
0.8 |
GO:0019227 |
neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 |
0.6 |
GO:0031848 |
protection from non-homologous end joining at telomere(GO:0031848) |
| 0.1 |
2.9 |
GO:0000038 |
very long-chain fatty acid metabolic process(GO:0000038) |
| 0.1 |
1.2 |
GO:0045792 |
negative regulation of cell size(GO:0045792) |
| 0.1 |
0.5 |
GO:0002002 |
regulation of angiotensin levels in blood(GO:0002002) |
| 0.1 |
1.7 |
GO:0001921 |
positive regulation of receptor recycling(GO:0001921) |
| 0.1 |
0.3 |
GO:0046726 |
positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.1 |
0.1 |
GO:0032661 |
regulation of interleukin-18 production(GO:0032661) negative regulation of interleukin-18 production(GO:0032701) |
| 0.1 |
0.5 |
GO:1904217 |
regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.1 |
0.4 |
GO:0034379 |
very-low-density lipoprotein particle assembly(GO:0034379) |
| 0.1 |
0.6 |
GO:0060662 |
tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.1 |
0.4 |
GO:0070814 |
hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.1 |
0.7 |
GO:0021860 |
pyramidal neuron development(GO:0021860) |
| 0.1 |
0.7 |
GO:0006817 |
phosphate ion transport(GO:0006817) |
| 0.1 |
0.4 |
GO:0061042 |
vascular wound healing(GO:0061042) |
| 0.1 |
0.3 |
GO:0071476 |
cellular hypotonic response(GO:0071476) |
| 0.1 |
0.5 |
GO:2000401 |
regulation of lymphocyte migration(GO:2000401) |
| 0.1 |
0.2 |
GO:0003430 |
growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.1 |
1.4 |
GO:0035458 |
cellular response to interferon-beta(GO:0035458) |
| 0.1 |
0.4 |
GO:0007220 |
Notch receptor processing(GO:0007220) membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 |
2.6 |
GO:0007157 |
heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 |
0.3 |
GO:1900170 |
negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.1 |
0.4 |
GO:0050882 |
voluntary musculoskeletal movement(GO:0050882) |
| 0.1 |
0.8 |
GO:0060218 |
hematopoietic stem cell differentiation(GO:0060218) |
| 0.1 |
0.5 |
GO:0010606 |
positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.1 |
0.1 |
GO:0010637 |
negative regulation of mitochondrial fusion(GO:0010637) |
| 0.1 |
0.4 |
GO:0000395 |
mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 |
0.3 |
GO:0042730 |
fibrinolysis(GO:0042730) |
| 0.1 |
1.2 |
GO:0033622 |
integrin activation(GO:0033622) |
| 0.1 |
1.3 |
GO:0032011 |
ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 |
0.2 |
GO:0008612 |
peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.0 |
0.7 |
GO:0035455 |
response to interferon-alpha(GO:0035455) |
| 0.0 |
0.3 |
GO:1903071 |
positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 |
0.1 |
GO:0035696 |
monocyte extravasation(GO:0035696) regulation of monocyte extravasation(GO:2000437) |
| 0.0 |
1.2 |
GO:0060004 |
reflex(GO:0060004) |
| 0.0 |
0.3 |
GO:0031665 |
negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 |
0.8 |
GO:0070884 |
regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 |
0.3 |
GO:0090383 |
phagosome acidification(GO:0090383) |
| 0.0 |
0.4 |
GO:0046415 |
urate metabolic process(GO:0046415) |
| 0.0 |
1.0 |
GO:0002091 |
negative regulation of receptor internalization(GO:0002091) |
| 0.0 |
0.6 |
GO:0018149 |
peptide cross-linking(GO:0018149) |
| 0.0 |
0.2 |
GO:0030854 |
positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 |
0.3 |
GO:0048680 |
positive regulation of axon regeneration(GO:0048680) |
| 0.0 |
0.3 |
GO:0051534 |
negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 |
1.6 |
GO:0071300 |
cellular response to retinoic acid(GO:0071300) |
| 0.0 |
0.3 |
GO:0071493 |
elastic fiber assembly(GO:0048251) cellular response to UV-B(GO:0071493) |
| 0.0 |
0.5 |
GO:0043407 |
negative regulation of MAP kinase activity(GO:0043407) |
| 0.0 |
0.4 |
GO:0032482 |
Rab protein signal transduction(GO:0032482) |
| 0.0 |
1.0 |
GO:0035162 |
embryonic hemopoiesis(GO:0035162) |
| 0.0 |
0.1 |
GO:0006990 |
positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.0 |
1.3 |
GO:0051567 |
histone H3-K9 methylation(GO:0051567) |
| 0.0 |
0.8 |
GO:0010107 |
potassium ion import(GO:0010107) |
| 0.0 |
0.5 |
GO:0006903 |
vesicle targeting(GO:0006903) |
| 0.0 |
0.4 |
GO:0032516 |
positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 |
0.4 |
GO:0097094 |
craniofacial suture morphogenesis(GO:0097094) |
| 0.0 |
0.2 |
GO:0035509 |
negative regulation of myosin-light-chain-phosphatase activity(GO:0035509) |
| 0.0 |
0.6 |
GO:0035066 |
positive regulation of histone acetylation(GO:0035066) |
| 0.0 |
0.1 |
GO:2001268 |
negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.0 |
0.2 |
GO:0017196 |
N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 |
0.3 |
GO:0019511 |
peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 |
0.3 |
GO:0034498 |
early endosome to Golgi transport(GO:0034498) |
| 0.0 |
0.2 |
GO:0019336 |
phenol-containing compound catabolic process(GO:0019336) |
| 0.0 |
1.5 |
GO:0003281 |
ventricular septum development(GO:0003281) |
| 0.0 |
2.4 |
GO:0002230 |
positive regulation of defense response to virus by host(GO:0002230) |
| 0.0 |
0.4 |
GO:0045907 |
positive regulation of vasoconstriction(GO:0045907) |
| 0.0 |
0.4 |
GO:0006182 |
cGMP biosynthetic process(GO:0006182) |
| 0.0 |
0.4 |
GO:0045652 |
regulation of megakaryocyte differentiation(GO:0045652) |
| 0.0 |
0.4 |
GO:0038092 |
nodal signaling pathway(GO:0038092) |
| 0.0 |
1.3 |
GO:0021766 |
hippocampus development(GO:0021766) |
| 0.0 |
0.7 |
GO:0006376 |
mRNA splice site selection(GO:0006376) |
| 0.0 |
0.6 |
GO:0046854 |
phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 |
1.0 |
GO:0016525 |
negative regulation of angiogenesis(GO:0016525) |
| 0.0 |
0.4 |
GO:0051457 |
maintenance of protein location in nucleus(GO:0051457) |
| 0.0 |
0.1 |
GO:0006977 |
DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) signal transduction involved in mitotic G1 DNA damage checkpoint(GO:0072431) intracellular signal transduction involved in G1 DNA damage checkpoint(GO:1902400) |
| 0.0 |
1.0 |
GO:0031122 |
cytoplasmic microtubule organization(GO:0031122) |
| 0.0 |
0.2 |
GO:0000479 |
endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.0 |
0.5 |
GO:0043666 |
regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 |
0.2 |
GO:0042554 |
superoxide anion generation(GO:0042554) |
| 0.0 |
0.3 |
GO:0071218 |
cellular response to misfolded protein(GO:0071218) |
| 0.0 |
1.5 |
GO:0090630 |
activation of GTPase activity(GO:0090630) |
| 0.0 |
0.4 |
GO:0046513 |
ceramide biosynthetic process(GO:0046513) |
| 0.0 |
0.5 |
GO:0051973 |
positive regulation of telomerase activity(GO:0051973) |
| 0.0 |
0.2 |
GO:0051382 |
kinetochore assembly(GO:0051382) |
| 0.0 |
0.7 |
GO:0030835 |
negative regulation of actin filament depolymerization(GO:0030835) |
| 0.0 |
0.1 |
GO:0016559 |
peroxisome fission(GO:0016559) |
| 0.0 |
0.5 |
GO:0034605 |
cellular response to heat(GO:0034605) |
| 0.0 |
0.3 |
GO:0042345 |
regulation of NF-kappaB import into nucleus(GO:0042345) NF-kappaB import into nucleus(GO:0042348) |
| 0.0 |
0.1 |
GO:0006560 |
proline metabolic process(GO:0006560) |
| 0.0 |
1.5 |
GO:0098656 |
anion transmembrane transport(GO:0098656) |
| 0.0 |
0.0 |
GO:0010724 |
regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 |
0.2 |
GO:0048268 |
clathrin coat assembly(GO:0048268) |
| 0.0 |
0.1 |
GO:0048096 |
chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 |
1.4 |
GO:0016579 |
protein deubiquitination(GO:0016579) |
| 0.0 |
0.2 |
GO:0006388 |
tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 |
0.2 |
GO:0010640 |
regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) |
| 0.0 |
2.0 |
GO:0043062 |
extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 |
0.1 |
GO:0006741 |
NADP biosynthetic process(GO:0006741) |
| 0.0 |
2.1 |
GO:0008654 |
phospholipid biosynthetic process(GO:0008654) |
| 0.0 |
0.6 |
GO:0006418 |
tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 |
0.2 |
GO:0071157 |
negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 |
0.6 |
GO:0006664 |
glycolipid metabolic process(GO:0006664) liposaccharide metabolic process(GO:1903509) |
| 0.0 |
0.2 |
GO:0007095 |
mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 |
0.3 |
GO:1903146 |
regulation of mitophagy(GO:1903146) |
| 0.0 |
0.4 |
GO:0051489 |
regulation of filopodium assembly(GO:0051489) |
| 0.0 |
0.2 |
GO:0006750 |
glutathione biosynthetic process(GO:0006750) |
| 0.0 |
0.6 |
GO:0000423 |
macromitophagy(GO:0000423) mitophagy in response to mitochondrial depolarization(GO:0098779) response to mitochondrial depolarisation(GO:0098780) |
| 0.0 |
0.1 |
GO:0046697 |
decidualization(GO:0046697) |
| 0.0 |
0.1 |
GO:0006102 |
isocitrate metabolic process(GO:0006102) |
| 0.0 |
0.5 |
GO:0006998 |
nuclear envelope organization(GO:0006998) |
| 0.0 |
0.1 |
GO:0008210 |
estrogen metabolic process(GO:0008210) |
| 0.0 |
0.0 |
GO:0071361 |
cellular response to ethanol(GO:0071361) |