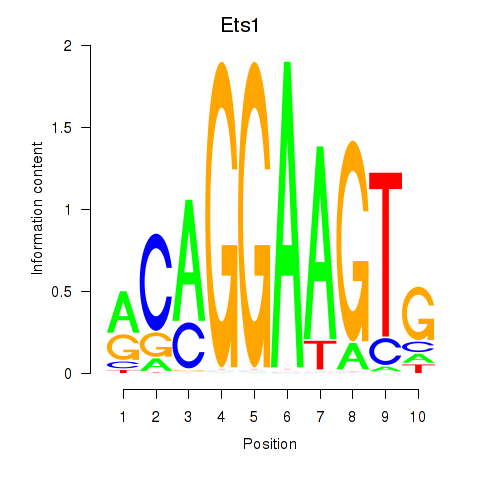
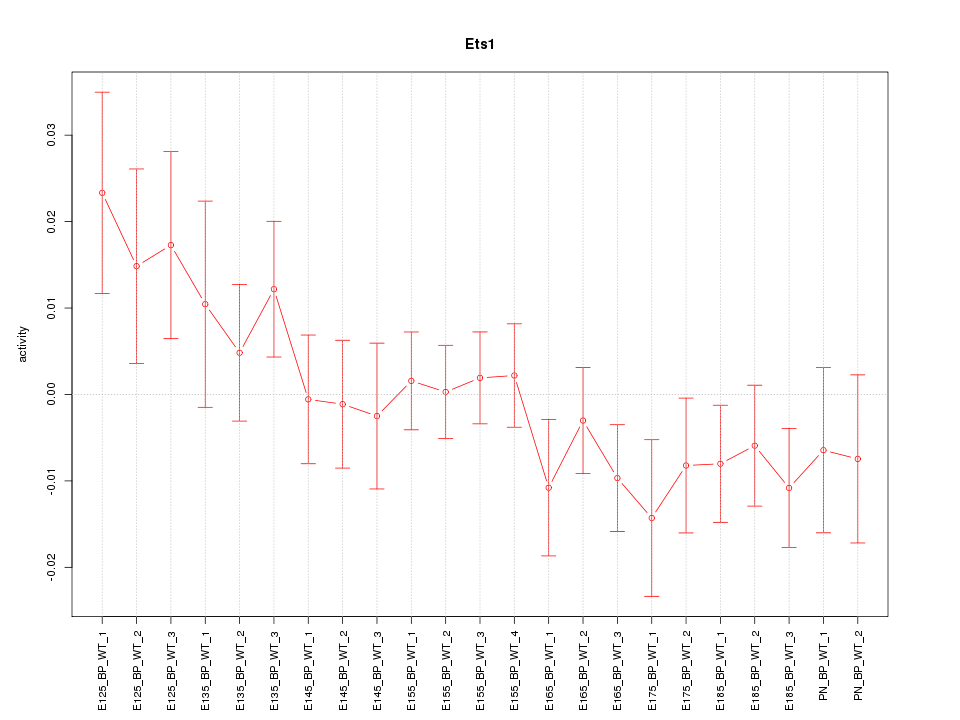
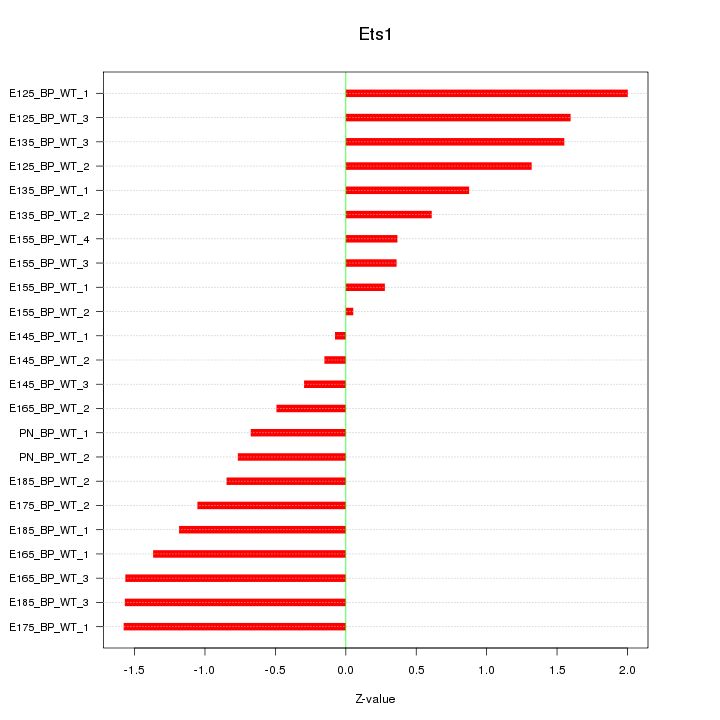
| 2.5 |
10.0 |
GO:0007056 |
spindle assembly involved in female meiosis(GO:0007056) |
| 1.2 |
3.7 |
GO:0016344 |
meiotic chromosome movement towards spindle pole(GO:0016344) |
| 1.2 |
3.7 |
GO:1901097 |
negative regulation of autophagosome maturation(GO:1901097) |
| 0.8 |
3.3 |
GO:0030576 |
Cajal body organization(GO:0030576) |
| 0.7 |
2.1 |
GO:1903896 |
positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.7 |
5.3 |
GO:0070474 |
positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.6 |
0.6 |
GO:0042713 |
sperm ejaculation(GO:0042713) |
| 0.6 |
1.8 |
GO:0021649 |
vestibulocochlear nerve morphogenesis(GO:0021648) vestibulocochlear nerve structural organization(GO:0021649) ganglion morphogenesis(GO:0061552) dorsal root ganglion morphogenesis(GO:1904835) |
| 0.6 |
3.0 |
GO:0043314 |
negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.6 |
1.7 |
GO:0060010 |
Sertoli cell fate commitment(GO:0060010) |
| 0.5 |
3.2 |
GO:0060666 |
dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.5 |
1.6 |
GO:1901552 |
positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.5 |
2.0 |
GO:0072385 |
minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.5 |
1.4 |
GO:0071921 |
establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.4 |
2.7 |
GO:0006452 |
translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.4 |
1.3 |
GO:0070889 |
platelet alpha granule organization(GO:0070889) |
| 0.4 |
1.2 |
GO:0001928 |
regulation of exocyst assembly(GO:0001928) |
| 0.4 |
1.2 |
GO:0015712 |
hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.4 |
1.5 |
GO:0046836 |
glycolipid transport(GO:0046836) |
| 0.4 |
1.5 |
GO:0051866 |
general adaptation syndrome(GO:0051866) |
| 0.4 |
1.4 |
GO:1903378 |
positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.4 |
1.1 |
GO:0032696 |
negative regulation of interleukin-13 production(GO:0032696) trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) odontoblast differentiation(GO:0071895) regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 0.4 |
1.4 |
GO:0060715 |
syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.3 |
0.7 |
GO:0010847 |
regulation of chromatin assembly(GO:0010847) |
| 0.3 |
1.4 |
GO:0061724 |
lipophagy(GO:0061724) |
| 0.3 |
1.3 |
GO:1902276 |
pancreatic amylase secretion(GO:0036395) regulation of pancreatic amylase secretion(GO:1902276) |
| 0.3 |
1.0 |
GO:0060399 |
positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.3 |
1.6 |
GO:0051490 |
negative regulation of filopodium assembly(GO:0051490) |
| 0.3 |
3.2 |
GO:0042989 |
sequestering of actin monomers(GO:0042989) |
| 0.3 |
2.2 |
GO:0051461 |
protein import into peroxisome matrix, docking(GO:0016560) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.3 |
0.9 |
GO:0002669 |
positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.3 |
1.5 |
GO:0007386 |
compartment pattern specification(GO:0007386) |
| 0.3 |
1.2 |
GO:0019355 |
nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.3 |
1.2 |
GO:1903898 |
negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.3 |
0.9 |
GO:0019405 |
alditol catabolic process(GO:0019405) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.3 |
0.9 |
GO:0042275 |
error-free postreplication DNA repair(GO:0042275) |
| 0.3 |
2.5 |
GO:0071493 |
cellular response to UV-B(GO:0071493) |
| 0.3 |
0.3 |
GO:1901187 |
regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.3 |
1.4 |
GO:1900017 |
positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.3 |
0.8 |
GO:0001830 |
trophectodermal cell fate commitment(GO:0001830) |
| 0.3 |
4.0 |
GO:0032927 |
positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.3 |
0.5 |
GO:1904457 |
positive regulation of neuronal action potential(GO:1904457) |
| 0.3 |
1.5 |
GO:0098734 |
protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.3 |
1.3 |
GO:1903772 |
regulation of viral budding via host ESCRT complex(GO:1903772) |
| 0.3 |
1.3 |
GO:0050651 |
dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.3 |
1.0 |
GO:0070537 |
histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.2 |
0.2 |
GO:1903899 |
positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.2 |
1.0 |
GO:0018343 |
protein farnesylation(GO:0018343) |
| 0.2 |
0.7 |
GO:0010499 |
proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.2 |
2.4 |
GO:0008063 |
Toll signaling pathway(GO:0008063) |
| 0.2 |
0.7 |
GO:0035790 |
platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.2 |
0.5 |
GO:1903599 |
positive regulation of mitophagy(GO:1903599) |
| 0.2 |
0.7 |
GO:0045472 |
response to ether(GO:0045472) |
| 0.2 |
0.5 |
GO:1902263 |
apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.2 |
1.3 |
GO:0010917 |
negative regulation of mitochondrial membrane potential(GO:0010917) |
| 0.2 |
0.7 |
GO:0006657 |
CDP-choline pathway(GO:0006657) |
| 0.2 |
0.6 |
GO:0036499 |
PERK-mediated unfolded protein response(GO:0036499) |
| 0.2 |
0.4 |
GO:0042997 |
negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.2 |
0.6 |
GO:0002014 |
vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.2 |
1.3 |
GO:0001771 |
immunological synapse formation(GO:0001771) |
| 0.2 |
1.1 |
GO:1900095 |
regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.2 |
0.6 |
GO:0070315 |
G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.2 |
0.4 |
GO:0031145 |
anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.2 |
0.2 |
GO:0090611 |
ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.2 |
0.6 |
GO:0070213 |
protein auto-ADP-ribosylation(GO:0070213) |
| 0.2 |
0.6 |
GO:1901228 |
positive regulation of skeletal muscle tissue growth(GO:0048633) positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 0.2 |
0.8 |
GO:0006742 |
NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.2 |
0.6 |
GO:0046368 |
GDP-L-fucose metabolic process(GO:0046368) |
| 0.2 |
3.4 |
GO:0051988 |
regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.2 |
1.0 |
GO:0002457 |
T cell antigen processing and presentation(GO:0002457) |
| 0.2 |
1.0 |
GO:0006348 |
chromatin silencing at telomere(GO:0006348) |
| 0.2 |
0.2 |
GO:0071638 |
negative regulation of chemokine production(GO:0032682) negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.2 |
1.3 |
GO:0000022 |
mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.2 |
0.6 |
GO:1901896 |
positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.2 |
3.6 |
GO:0060236 |
regulation of mitotic spindle organization(GO:0060236) |
| 0.2 |
1.1 |
GO:0035743 |
CD4-positive, alpha-beta T cell cytokine production(GO:0035743) T-helper 2 cell cytokine production(GO:0035745) |
| 0.2 |
1.9 |
GO:0002693 |
positive regulation of cellular extravasation(GO:0002693) |
| 0.2 |
1.1 |
GO:1990564 |
protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.2 |
0.7 |
GO:0044565 |
dendritic cell proliferation(GO:0044565) |
| 0.2 |
0.6 |
GO:0060743 |
epithelial cell maturation involved in prostate gland development(GO:0060743) alveolar secondary septum development(GO:0061144) |
| 0.2 |
1.5 |
GO:1900262 |
regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.2 |
0.7 |
GO:0010735 |
positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.2 |
0.4 |
GO:1900227 |
positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.2 |
0.9 |
GO:0046016 |
regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) positive regulation of transcription by glucose(GO:0046016) |
| 0.2 |
0.9 |
GO:0001842 |
neural fold formation(GO:0001842) |
| 0.2 |
1.1 |
GO:0070863 |
positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.2 |
0.5 |
GO:2000393 |
negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.2 |
1.2 |
GO:1901977 |
negative regulation of cell cycle checkpoint(GO:1901977) negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.2 |
0.4 |
GO:1902915 |
negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.2 |
0.9 |
GO:1903361 |
protein localization to basolateral plasma membrane(GO:1903361) |
| 0.2 |
0.5 |
GO:1902474 |
positive regulation of protein localization to synapse(GO:1902474) |
| 0.2 |
1.9 |
GO:0097034 |
mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.2 |
0.9 |
GO:0034196 |
acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.2 |
0.5 |
GO:0061588 |
calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylserine scrambling(GO:0061589) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.2 |
0.3 |
GO:0031914 |
negative regulation of synaptic plasticity(GO:0031914) |
| 0.2 |
0.5 |
GO:1904211 |
membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.2 |
0.7 |
GO:0046898 |
response to cycloheximide(GO:0046898) |
| 0.2 |
0.5 |
GO:0075525 |
viral translational termination-reinitiation(GO:0075525) |
| 0.2 |
1.1 |
GO:1903715 |
regulation of aerobic respiration(GO:1903715) |
| 0.2 |
1.8 |
GO:0045899 |
positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.2 |
3.0 |
GO:0061000 |
negative regulation of dendritic spine development(GO:0061000) |
| 0.2 |
0.6 |
GO:0007258 |
JUN phosphorylation(GO:0007258) |
| 0.2 |
0.2 |
GO:2000574 |
regulation of microtubule motor activity(GO:2000574) |
| 0.2 |
1.1 |
GO:0007213 |
G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.2 |
0.9 |
GO:0045213 |
neurotransmitter receptor metabolic process(GO:0045213) |
| 0.2 |
1.2 |
GO:0036506 |
maintenance of unfolded protein(GO:0036506) protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.2 |
2.9 |
GO:0007413 |
axonal fasciculation(GO:0007413) |
| 0.1 |
0.4 |
GO:0071169 |
establishment of protein localization to chromatin(GO:0071169) |
| 0.1 |
0.1 |
GO:0032627 |
interleukin-23 production(GO:0032627) regulation of interleukin-23 production(GO:0032667) |
| 0.1 |
1.5 |
GO:1903504 |
regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.1 |
0.1 |
GO:0032829 |
regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) |
| 0.1 |
0.7 |
GO:0042637 |
catagen(GO:0042637) |
| 0.1 |
1.3 |
GO:0098907 |
protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) regulation of SA node cell action potential(GO:0098907) |
| 0.1 |
0.9 |
GO:0006123 |
mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 |
2.3 |
GO:0043248 |
proteasome assembly(GO:0043248) |
| 0.1 |
0.4 |
GO:0070476 |
rRNA (guanine-N7)-methylation(GO:0070476) |
| 0.1 |
1.2 |
GO:0033182 |
regulation of histone ubiquitination(GO:0033182) |
| 0.1 |
0.4 |
GO:1900126 |
negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.1 |
0.4 |
GO:0031424 |
keratinization(GO:0031424) |
| 0.1 |
0.4 |
GO:0060744 |
thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.1 |
0.3 |
GO:0035021 |
negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.1 |
2.5 |
GO:0045116 |
protein neddylation(GO:0045116) |
| 0.1 |
1.0 |
GO:0061158 |
3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.1 |
2.1 |
GO:0071801 |
regulation of podosome assembly(GO:0071801) |
| 0.1 |
0.3 |
GO:0080009 |
mRNA methylation(GO:0080009) |
| 0.1 |
0.3 |
GO:0070944 |
neutrophil mediated killing of bacterium(GO:0070944) |
| 0.1 |
0.3 |
GO:0090135 |
actin filament branching(GO:0090135) |
| 0.1 |
0.8 |
GO:0071883 |
activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.1 |
0.4 |
GO:0051030 |
snRNA transport(GO:0051030) |
| 0.1 |
0.8 |
GO:0007197 |
adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 |
0.4 |
GO:0002322 |
B cell proliferation involved in immune response(GO:0002322) |
| 0.1 |
0.9 |
GO:2000286 |
receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.1 |
0.6 |
GO:0090164 |
asymmetric Golgi ribbon formation(GO:0090164) |
| 0.1 |
1.5 |
GO:0032506 |
cytokinetic process(GO:0032506) |
| 0.1 |
2.5 |
GO:0016226 |
iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 |
0.2 |
GO:0002069 |
columnar/cuboidal epithelial cell maturation(GO:0002069) |
| 0.1 |
0.5 |
GO:0021812 |
neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.1 |
0.5 |
GO:0071033 |
nuclear retention of pre-mRNA at the site of transcription(GO:0071033) CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.1 |
2.4 |
GO:0032968 |
positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.1 |
0.2 |
GO:0002309 |
T cell proliferation involved in immune response(GO:0002309) |
| 0.1 |
3.4 |
GO:0010960 |
magnesium ion homeostasis(GO:0010960) |
| 0.1 |
0.6 |
GO:0046543 |
development of secondary female sexual characteristics(GO:0046543) |
| 0.1 |
0.5 |
GO:0042905 |
9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 |
0.4 |
GO:0021691 |
cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.1 |
2.6 |
GO:0010763 |
positive regulation of fibroblast migration(GO:0010763) |
| 0.1 |
0.6 |
GO:0031052 |
programmed DNA elimination(GO:0031049) chromosome breakage(GO:0031052) |
| 0.1 |
0.5 |
GO:0034421 |
post-translational protein acetylation(GO:0034421) |
| 0.1 |
0.5 |
GO:1902626 |
assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 |
0.3 |
GO:1900122 |
positive regulation of receptor binding(GO:1900122) |
| 0.1 |
0.8 |
GO:0030836 |
positive regulation of actin filament depolymerization(GO:0030836) |
| 0.1 |
0.3 |
GO:0032241 |
positive regulation of nucleobase-containing compound transport(GO:0032241) |
| 0.1 |
0.3 |
GO:0060124 |
positive regulation of growth hormone secretion(GO:0060124) |
| 0.1 |
0.2 |
GO:0070560 |
protein secretion by platelet(GO:0070560) |
| 0.1 |
0.4 |
GO:0072344 |
rescue of stalled ribosome(GO:0072344) |
| 0.1 |
0.8 |
GO:0051014 |
actin filament severing(GO:0051014) |
| 0.1 |
0.5 |
GO:2000189 |
positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.1 |
0.4 |
GO:0009052 |
pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.1 |
0.2 |
GO:0090232 |
positive regulation of spindle checkpoint(GO:0090232) |
| 0.1 |
0.4 |
GO:0033314 |
mitotic DNA replication checkpoint(GO:0033314) |
| 0.1 |
0.3 |
GO:0002432 |
granuloma formation(GO:0002432) |
| 0.1 |
0.7 |
GO:0033601 |
positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.1 |
1.1 |
GO:0060837 |
blood vessel endothelial cell differentiation(GO:0060837) |
| 0.1 |
0.8 |
GO:0070317 |
negative regulation of G0 to G1 transition(GO:0070317) |
| 0.1 |
0.3 |
GO:0090091 |
positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.1 |
1.1 |
GO:0006968 |
cellular defense response(GO:0006968) |
| 0.1 |
0.7 |
GO:0051418 |
interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.1 |
3.7 |
GO:0000387 |
spliceosomal snRNP assembly(GO:0000387) |
| 0.1 |
0.4 |
GO:0001550 |
ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 |
0.6 |
GO:1904263 |
positive regulation of TORC1 signaling(GO:1904263) |
| 0.1 |
0.9 |
GO:0032464 |
positive regulation of protein homooligomerization(GO:0032464) |
| 0.1 |
0.3 |
GO:0001732 |
formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.1 |
1.1 |
GO:0061179 |
negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 |
1.3 |
GO:0001731 |
formation of translation preinitiation complex(GO:0001731) |
| 0.1 |
0.3 |
GO:1904690 |
regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.1 |
0.3 |
GO:2000834 |
androgen secretion(GO:0035935) testosterone secretion(GO:0035936) regulation of androgen secretion(GO:2000834) positive regulation of androgen secretion(GO:2000836) regulation of testosterone secretion(GO:2000843) positive regulation of testosterone secretion(GO:2000845) |
| 0.1 |
0.2 |
GO:0038161 |
prolactin signaling pathway(GO:0038161) |
| 0.1 |
0.3 |
GO:0006597 |
spermine biosynthetic process(GO:0006597) |
| 0.1 |
0.3 |
GO:0032511 |
late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 |
1.4 |
GO:0009191 |
ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.1 |
1.2 |
GO:0006654 |
phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.1 |
1.0 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.1 |
2.4 |
GO:0030970 |
retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.1 |
0.2 |
GO:2000152 |
regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) regulation of ubiquitin-specific protease activity(GO:2000152) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.1 |
0.3 |
GO:2000508 |
regulation of dendritic cell chemotaxis(GO:2000508) positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.1 |
0.2 |
GO:0070166 |
enamel mineralization(GO:0070166) |
| 0.1 |
1.4 |
GO:2000251 |
positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 |
0.4 |
GO:0036336 |
dendritic cell migration(GO:0036336) |
| 0.1 |
1.6 |
GO:0010569 |
regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.1 |
1.2 |
GO:0031573 |
intra-S DNA damage checkpoint(GO:0031573) |
| 0.1 |
0.3 |
GO:0070459 |
prolactin secretion(GO:0070459) |
| 0.1 |
0.8 |
GO:0046036 |
GTP biosynthetic process(GO:0006183) CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.1 |
0.6 |
GO:0009048 |
dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.1 |
0.5 |
GO:0032308 |
positive regulation of prostaglandin secretion(GO:0032308) |
| 0.1 |
0.4 |
GO:0060279 |
positive regulation of ovulation(GO:0060279) |
| 0.1 |
0.2 |
GO:0070345 |
negative regulation of fat cell proliferation(GO:0070345) |
| 0.1 |
0.4 |
GO:0030259 |
lipid glycosylation(GO:0030259) |
| 0.1 |
0.6 |
GO:0006297 |
nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 |
0.2 |
GO:1903232 |
melanosome assembly(GO:1903232) |
| 0.1 |
0.3 |
GO:0032020 |
ISG15-protein conjugation(GO:0032020) |
| 0.1 |
1.1 |
GO:0043249 |
erythrocyte maturation(GO:0043249) |
| 0.1 |
2.6 |
GO:0003301 |
physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 |
3.0 |
GO:0046825 |
regulation of protein export from nucleus(GO:0046825) |
| 0.1 |
0.5 |
GO:1901029 |
negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.1 |
0.2 |
GO:0009298 |
GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 |
0.7 |
GO:0033148 |
positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.1 |
0.8 |
GO:0048386 |
positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 |
0.2 |
GO:1901069 |
guanosine-containing compound catabolic process(GO:1901069) |
| 0.1 |
0.5 |
GO:0070836 |
caveola assembly(GO:0070836) |
| 0.1 |
0.4 |
GO:0046684 |
response to pyrethroid(GO:0046684) |
| 0.1 |
0.2 |
GO:0006420 |
arginyl-tRNA aminoacylation(GO:0006420) |
| 0.1 |
0.2 |
GO:0006269 |
DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 |
1.1 |
GO:0036260 |
7-methylguanosine RNA capping(GO:0009452) RNA capping(GO:0036260) |
| 0.1 |
0.3 |
GO:1903031 |
regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.1 |
0.4 |
GO:0060046 |
regulation of acrosome reaction(GO:0060046) |
| 0.1 |
2.5 |
GO:0070979 |
protein K11-linked ubiquitination(GO:0070979) |
| 0.1 |
1.9 |
GO:0045722 |
positive regulation of gluconeogenesis(GO:0045722) |
| 0.1 |
0.9 |
GO:0090141 |
positive regulation of mitochondrial fission(GO:0090141) |
| 0.1 |
0.4 |
GO:0045218 |
zonula adherens maintenance(GO:0045218) |
| 0.1 |
1.0 |
GO:0032060 |
bleb assembly(GO:0032060) |
| 0.1 |
0.7 |
GO:0043552 |
positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.1 |
0.3 |
GO:0019372 |
lipoxygenase pathway(GO:0019372) |
| 0.1 |
0.3 |
GO:0009142 |
nucleoside triphosphate biosynthetic process(GO:0009142) |
| 0.1 |
0.5 |
GO:0071318 |
cellular response to ATP(GO:0071318) |
| 0.1 |
0.1 |
GO:0010871 |
negative regulation of receptor biosynthetic process(GO:0010871) |
| 0.1 |
0.3 |
GO:2000672 |
negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.1 |
0.2 |
GO:0006436 |
tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 |
0.5 |
GO:0007296 |
vitellogenesis(GO:0007296) |
| 0.1 |
0.5 |
GO:0032625 |
interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 |
0.6 |
GO:1902018 |
negative regulation of cilium assembly(GO:1902018) |
| 0.1 |
0.5 |
GO:0070935 |
3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.1 |
0.8 |
GO:1900383 |
regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 |
3.2 |
GO:0043484 |
regulation of RNA splicing(GO:0043484) |
| 0.1 |
0.6 |
GO:1903608 |
protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.1 |
0.5 |
GO:0040016 |
embryonic cleavage(GO:0040016) |
| 0.1 |
0.4 |
GO:2000491 |
positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.1 |
0.3 |
GO:0010626 |
negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.1 |
0.8 |
GO:0010592 |
positive regulation of lamellipodium assembly(GO:0010592) |
| 0.1 |
0.6 |
GO:0018065 |
protein-cofactor linkage(GO:0018065) |
| 0.1 |
0.3 |
GO:0015786 |
UDP-glucose transport(GO:0015786) |
| 0.1 |
2.6 |
GO:0006284 |
base-excision repair(GO:0006284) |
| 0.1 |
0.5 |
GO:0035735 |
intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 |
0.4 |
GO:0007096 |
regulation of exit from mitosis(GO:0007096) |
| 0.1 |
0.7 |
GO:0046628 |
positive regulation of insulin receptor signaling pathway(GO:0046628) |
| 0.1 |
0.4 |
GO:0008053 |
mitochondrial fusion(GO:0008053) |
| 0.1 |
0.2 |
GO:0045650 |
negative regulation of macrophage differentiation(GO:0045650) |
| 0.1 |
0.1 |
GO:0044650 |
virion attachment to host cell(GO:0019062) adhesion of symbiont to host cell(GO:0044650) |
| 0.1 |
0.2 |
GO:0000389 |
mRNA 3'-splice site recognition(GO:0000389) |
| 0.1 |
0.4 |
GO:0018342 |
protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.1 |
0.2 |
GO:0006379 |
mRNA cleavage(GO:0006379) |
| 0.1 |
2.4 |
GO:0010501 |
RNA secondary structure unwinding(GO:0010501) |
| 0.1 |
0.6 |
GO:0045040 |
protein import into mitochondrial outer membrane(GO:0045040) |
| 0.1 |
0.5 |
GO:0050774 |
negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.1 |
2.0 |
GO:0006378 |
mRNA polyadenylation(GO:0006378) |
| 0.1 |
0.2 |
GO:1902969 |
mitotic DNA replication(GO:1902969) |
| 0.1 |
0.5 |
GO:0000083 |
regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 |
0.4 |
GO:0048742 |
regulation of skeletal muscle fiber development(GO:0048742) |
| 0.1 |
0.4 |
GO:0000012 |
single strand break repair(GO:0000012) |
| 0.1 |
2.5 |
GO:0045761 |
regulation of adenylate cyclase activity(GO:0045761) |
| 0.1 |
0.4 |
GO:0032486 |
Rap protein signal transduction(GO:0032486) |
| 0.1 |
0.2 |
GO:0031509 |
telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.1 |
0.6 |
GO:0039703 |
viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.1 |
0.3 |
GO:0033632 |
regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.1 |
0.9 |
GO:0043968 |
histone H2A acetylation(GO:0043968) |
| 0.1 |
0.2 |
GO:0035617 |
cytoplasm organization(GO:0007028) stress granule disassembly(GO:0035617) plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.1 |
0.3 |
GO:0008611 |
ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.1 |
0.2 |
GO:0042117 |
monocyte activation(GO:0042117) |
| 0.1 |
0.2 |
GO:0002949 |
tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 |
0.2 |
GO:1903236 |
regulation of leukocyte tethering or rolling(GO:1903236) negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.1 |
0.6 |
GO:0031268 |
pseudopodium organization(GO:0031268) |
| 0.1 |
0.1 |
GO:0046015 |
regulation of transcription by glucose(GO:0046015) |
| 0.1 |
0.2 |
GO:0006933 |
negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.1 |
1.2 |
GO:0006910 |
phagocytosis, recognition(GO:0006910) |
| 0.1 |
0.5 |
GO:0009396 |
folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.1 |
0.5 |
GO:0042492 |
gamma-delta T cell differentiation(GO:0042492) |
| 0.0 |
2.0 |
GO:0015804 |
neutral amino acid transport(GO:0015804) |
| 0.0 |
0.2 |
GO:0030242 |
pexophagy(GO:0030242) |
| 0.0 |
2.6 |
GO:0050830 |
defense response to Gram-positive bacterium(GO:0050830) |
| 0.0 |
0.2 |
GO:0035562 |
negative regulation of chromatin binding(GO:0035562) |
| 0.0 |
0.4 |
GO:2001223 |
negative regulation of neuron migration(GO:2001223) |
| 0.0 |
0.4 |
GO:0042832 |
response to protozoan(GO:0001562) defense response to protozoan(GO:0042832) |
| 0.0 |
0.5 |
GO:0006415 |
translational termination(GO:0006415) |
| 0.0 |
0.1 |
GO:0032066 |
nucleolus to nucleoplasm transport(GO:0032066) |
| 0.0 |
0.6 |
GO:0043984 |
histone H4-K16 acetylation(GO:0043984) |
| 0.0 |
0.8 |
GO:0006911 |
phagocytosis, engulfment(GO:0006911) |
| 0.0 |
0.7 |
GO:0048026 |
positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 |
0.7 |
GO:0043486 |
histone exchange(GO:0043486) |
| 0.0 |
0.3 |
GO:0035020 |
regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 |
0.4 |
GO:0006020 |
inositol metabolic process(GO:0006020) |
| 0.0 |
0.2 |
GO:0006432 |
phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 |
0.3 |
GO:0045974 |
miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 |
0.2 |
GO:0030263 |
apoptotic chromosome condensation(GO:0030263) |
| 0.0 |
1.5 |
GO:0072661 |
protein targeting to plasma membrane(GO:0072661) |
| 0.0 |
0.5 |
GO:1990173 |
protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.0 |
0.3 |
GO:0006398 |
mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 |
0.1 |
GO:0070172 |
positive regulation of tooth mineralization(GO:0070172) |
| 0.0 |
0.6 |
GO:0048387 |
negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 |
0.4 |
GO:0030388 |
fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 |
0.6 |
GO:0050765 |
negative regulation of phagocytosis(GO:0050765) |
| 0.0 |
0.3 |
GO:0010825 |
positive regulation of centrosome duplication(GO:0010825) |
| 0.0 |
2.4 |
GO:0022900 |
electron transport chain(GO:0022900) |
| 0.0 |
0.9 |
GO:0010800 |
positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 |
0.6 |
GO:0000077 |
DNA damage checkpoint(GO:0000077) |
| 0.0 |
0.4 |
GO:0035855 |
megakaryocyte development(GO:0035855) |
| 0.0 |
0.2 |
GO:0051697 |
protein delipidation(GO:0051697) |
| 0.0 |
0.6 |
GO:0032515 |
negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 |
0.6 |
GO:0070816 |
phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 |
0.8 |
GO:0050853 |
B cell receptor signaling pathway(GO:0050853) |
| 0.0 |
0.1 |
GO:0006990 |
positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.0 |
0.2 |
GO:0071361 |
cellular response to ethanol(GO:0071361) |
| 0.0 |
0.1 |
GO:0060800 |
regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 |
2.0 |
GO:0006367 |
transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 |
0.2 |
GO:0043353 |
enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 |
0.1 |
GO:1902950 |
regulation of dendritic spine maintenance(GO:1902950) positive regulation of dendritic spine maintenance(GO:1902952) |
| 0.0 |
0.1 |
GO:0046541 |
saliva secretion(GO:0046541) |
| 0.0 |
0.2 |
GO:0006995 |
cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 |
0.3 |
GO:0090043 |
regulation of tubulin deacetylation(GO:0090043) |
| 0.0 |
0.2 |
GO:0014022 |
neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 |
0.1 |
GO:0060546 |
negative regulation of necroptotic process(GO:0060546) |
| 0.0 |
1.0 |
GO:0043044 |
ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 |
6.8 |
GO:0000398 |
RNA splicing, via transesterification reactions(GO:0000375) RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 |
0.6 |
GO:2000279 |
negative regulation of DNA biosynthetic process(GO:2000279) |
| 0.0 |
0.1 |
GO:0036292 |
DNA rewinding(GO:0036292) |
| 0.0 |
0.6 |
GO:0006221 |
pyrimidine nucleotide biosynthetic process(GO:0006221) |
| 0.0 |
1.0 |
GO:0048488 |
synaptic vesicle endocytosis(GO:0048488) |
| 0.0 |
0.1 |
GO:1902455 |
negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 |
0.2 |
GO:0097369 |
sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 |
0.1 |
GO:0010757 |
arachidonic acid metabolite production involved in inflammatory response(GO:0002538) negative regulation of plasminogen activation(GO:0010757) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 |
0.2 |
GO:2000270 |
negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 |
0.3 |
GO:0010832 |
negative regulation of myotube differentiation(GO:0010832) |
| 0.0 |
0.3 |
GO:0045947 |
negative regulation of translational initiation(GO:0045947) |
| 0.0 |
0.1 |
GO:0070488 |
regulation of integrin biosynthetic process(GO:0045113) neutrophil aggregation(GO:0070488) |
| 0.0 |
0.3 |
GO:0032462 |
regulation of protein homooligomerization(GO:0032462) |
| 0.0 |
0.5 |
GO:0072583 |
clathrin-mediated endocytosis(GO:0072583) |
| 0.0 |
0.5 |
GO:0050684 |
regulation of mRNA processing(GO:0050684) |
| 0.0 |
0.3 |
GO:0090435 |
protein localization to nuclear envelope(GO:0090435) |
| 0.0 |
0.1 |
GO:0018242 |
protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 |
0.4 |
GO:0043001 |
Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 |
0.5 |
GO:0048268 |
clathrin coat assembly(GO:0048268) |
| 0.0 |
0.4 |
GO:0048875 |
chemical homeostasis within a tissue(GO:0048875) |
| 0.0 |
0.1 |
GO:0090201 |
negative regulation of release of cytochrome c from mitochondria(GO:0090201) |
| 0.0 |
0.1 |
GO:0018199 |
peptidyl-glutamine modification(GO:0018199) |
| 0.0 |
0.1 |
GO:0032000 |
positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 |
0.4 |
GO:0046677 |
response to antibiotic(GO:0046677) |
| 0.0 |
0.1 |
GO:0031125 |
rRNA 3'-end processing(GO:0031125) |
| 0.0 |
0.3 |
GO:0046855 |
phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 |
0.1 |
GO:1902897 |
regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.0 |
0.6 |
GO:0097150 |
neuronal stem cell population maintenance(GO:0097150) |
| 0.0 |
0.5 |
GO:2000311 |
regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 |
0.4 |
GO:0006744 |
ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 |
0.6 |
GO:0000154 |
rRNA modification(GO:0000154) |
| 0.0 |
0.1 |
GO:2000036 |
regulation of stem cell population maintenance(GO:2000036) |
| 0.0 |
0.4 |
GO:0045807 |
positive regulation of endocytosis(GO:0045807) |
| 0.0 |
1.7 |
GO:2001237 |
negative regulation of extrinsic apoptotic signaling pathway(GO:2001237) |
| 0.0 |
0.1 |
GO:0000469 |
cleavage involved in rRNA processing(GO:0000469) endonucleolytic cleavage involved in rRNA processing(GO:0000478) |
| 0.0 |
1.0 |
GO:0002181 |
cytoplasmic translation(GO:0002181) |
| 0.0 |
0.1 |
GO:1903587 |
regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 0.0 |
0.4 |
GO:0043550 |
regulation of lipid kinase activity(GO:0043550) |
| 0.0 |
0.2 |
GO:0000290 |
deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 |
0.2 |
GO:0010815 |
bradykinin catabolic process(GO:0010815) |
| 0.0 |
1.7 |
GO:0031398 |
positive regulation of protein ubiquitination(GO:0031398) |
| 0.0 |
0.1 |
GO:0035752 |
lysosomal lumen pH elevation(GO:0035752) |
| 0.0 |
0.2 |
GO:0042053 |
regulation of dopamine metabolic process(GO:0042053) |
| 0.0 |
1.0 |
GO:0042073 |
intraciliary transport(GO:0042073) |
| 0.0 |
0.1 |
GO:0090073 |
positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 |
0.1 |
GO:0034472 |
snRNA 3'-end processing(GO:0034472) |
| 0.0 |
0.4 |
GO:0010603 |
regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 |
0.4 |
GO:0034643 |
establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 |
0.2 |
GO:0032780 |
negative regulation of ATPase activity(GO:0032780) |
| 0.0 |
0.1 |
GO:0046061 |
dATP catabolic process(GO:0046061) |
| 0.0 |
2.4 |
GO:0010466 |
negative regulation of peptidase activity(GO:0010466) |
| 0.0 |
0.8 |
GO:0035329 |
hippo signaling(GO:0035329) |
| 0.0 |
1.3 |
GO:0042274 |
ribosomal small subunit biogenesis(GO:0042274) |
| 0.0 |
0.1 |
GO:0051096 |
regulation of helicase activity(GO:0051095) positive regulation of helicase activity(GO:0051096) |
| 0.0 |
0.3 |
GO:0043388 |
positive regulation of DNA binding(GO:0043388) |
| 0.0 |
0.2 |
GO:1902857 |
positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 |
0.2 |
GO:0060134 |
prepulse inhibition(GO:0060134) |
| 0.0 |
0.2 |
GO:0042759 |
long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 |
0.0 |
GO:0010635 |
regulation of mitochondrial fusion(GO:0010635) negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 |
0.2 |
GO:0035751 |
regulation of lysosomal lumen pH(GO:0035751) |
| 0.0 |
0.1 |
GO:0043201 |
response to leucine(GO:0043201) cellular response to leucine(GO:0071233) |
| 0.0 |
0.2 |
GO:2000310 |
regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.0 |
0.0 |
GO:2000402 |
negative regulation of lymphocyte migration(GO:2000402) |
| 0.0 |
0.1 |
GO:0001302 |
replicative cell aging(GO:0001302) |
| 0.0 |
0.7 |
GO:0010765 |
positive regulation of sodium ion transport(GO:0010765) |
| 0.0 |
0.3 |
GO:0099517 |
anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 |
0.6 |
GO:0010633 |
negative regulation of epithelial cell migration(GO:0010633) |
| 0.0 |
0.4 |
GO:0014003 |
oligodendrocyte development(GO:0014003) |
| 0.0 |
0.1 |
GO:0045197 |
establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 |
0.2 |
GO:0051043 |
regulation of membrane protein ectodomain proteolysis(GO:0051043) |
| 0.0 |
0.2 |
GO:0090036 |
regulation of protein kinase C signaling(GO:0090036) |
| 0.0 |
1.7 |
GO:0006626 |
protein targeting to mitochondrion(GO:0006626) |
| 0.0 |
0.1 |
GO:0061484 |
hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 |
0.0 |
GO:0001927 |
exocyst assembly(GO:0001927) |
| 0.0 |
0.0 |
GO:0090383 |
phagosome acidification(GO:0090383) |
| 0.0 |
0.2 |
GO:0010388 |
protein deneddylation(GO:0000338) cullin deneddylation(GO:0010388) |
| 0.0 |
0.1 |
GO:0030049 |
muscle filament sliding(GO:0030049) |
| 0.0 |
0.4 |
GO:0031146 |
SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 |
0.1 |
GO:0006406 |
mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 |
0.1 |
GO:0098535 |
de novo centriole assembly(GO:0098535) |
| 0.0 |
0.2 |
GO:0016180 |
snRNA processing(GO:0016180) |
| 0.0 |
0.2 |
GO:0006656 |
phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 |
0.2 |
GO:0046069 |
cGMP catabolic process(GO:0046069) |
| 0.0 |
0.4 |
GO:0034723 |
DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 |
0.1 |
GO:0061756 |
leukocyte tethering or rolling(GO:0050901) leukocyte adhesion to vascular endothelial cell(GO:0061756) |
| 0.0 |
0.5 |
GO:0055007 |
cardiac muscle cell differentiation(GO:0055007) |
| 0.0 |
0.1 |
GO:0031584 |
activation of phospholipase D activity(GO:0031584) |
| 0.0 |
0.9 |
GO:0030148 |
sphingolipid biosynthetic process(GO:0030148) |
| 0.0 |
0.1 |
GO:0051601 |
exocyst localization(GO:0051601) |
| 0.0 |
0.8 |
GO:0006888 |
ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 |
0.2 |
GO:0071108 |
protein K48-linked deubiquitination(GO:0071108) |
| 0.0 |
0.2 |
GO:1902850 |
mitotic spindle assembly(GO:0090307) microtubule cytoskeleton organization involved in mitosis(GO:1902850) |
| 0.0 |
0.2 |
GO:0007193 |
adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 |
0.1 |
GO:0050435 |
beta-amyloid metabolic process(GO:0050435) |
| 0.0 |
0.1 |
GO:1900078 |
positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 |
0.3 |
GO:0006383 |
transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 |
0.3 |
GO:0045746 |
negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 |
0.1 |
GO:0098927 |
early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.0 |
0.2 |
GO:0010669 |
epithelial structure maintenance(GO:0010669) |
| 0.0 |
0.4 |
GO:0007585 |
respiratory gaseous exchange(GO:0007585) |
| 0.0 |
0.2 |
GO:0001706 |
endoderm formation(GO:0001706) |