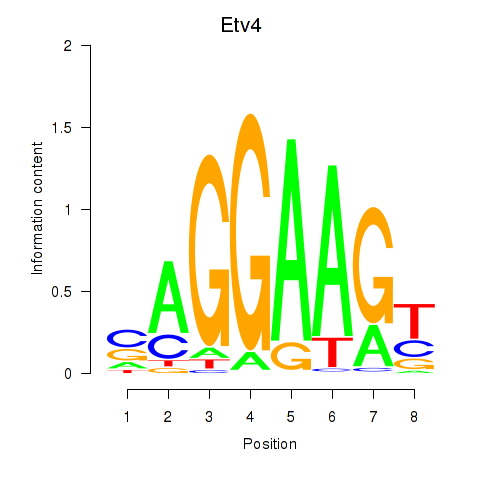
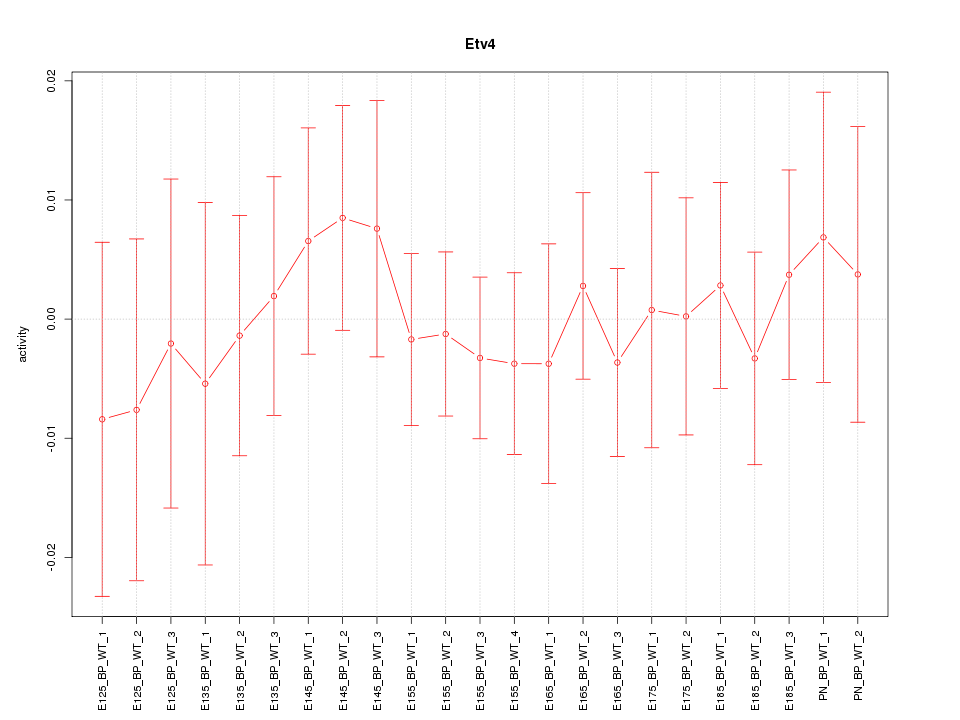
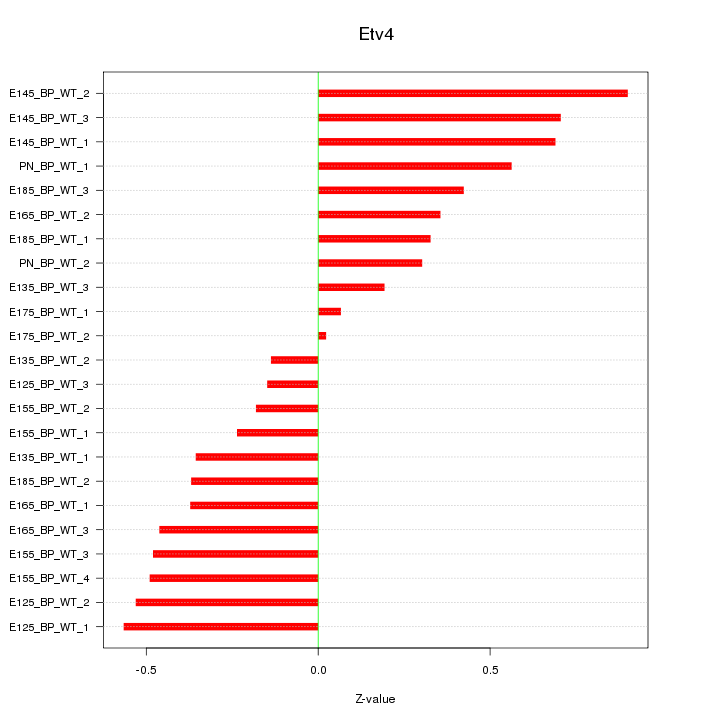
| 0.2 |
0.6 |
GO:0003032 |
detection of oxygen(GO:0003032) |
| 0.1 |
1.2 |
GO:0036506 |
maintenance of unfolded protein(GO:0036506) protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.1 |
0.5 |
GO:0021564 |
glossopharyngeal nerve development(GO:0021563) vagus nerve development(GO:0021564) |
| 0.1 |
0.3 |
GO:0048611 |
ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.1 |
0.3 |
GO:0036092 |
phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 |
0.3 |
GO:0043553 |
negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 |
1.0 |
GO:2001214 |
positive regulation of vasculogenesis(GO:2001214) |
| 0.1 |
0.5 |
GO:0070278 |
extracellular matrix constituent secretion(GO:0070278) |
| 0.1 |
0.3 |
GO:0032497 |
detection of lipopolysaccharide(GO:0032497) |
| 0.1 |
0.5 |
GO:0043654 |
recognition of apoptotic cell(GO:0043654) |
| 0.1 |
0.4 |
GO:0051189 |
Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.1 |
0.3 |
GO:0006624 |
vacuolar protein processing(GO:0006624) |
| 0.1 |
0.1 |
GO:1901668 |
regulation of superoxide dismutase activity(GO:1901668) |
| 0.1 |
0.8 |
GO:0090232 |
positive regulation of spindle checkpoint(GO:0090232) |
| 0.1 |
0.2 |
GO:0042939 |
renal sodium ion transport(GO:0003096) glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.1 |
0.1 |
GO:0032079 |
positive regulation of endodeoxyribonuclease activity(GO:0032079) |
| 0.1 |
0.3 |
GO:0014846 |
esophagus smooth muscle contraction(GO:0014846) |
| 0.1 |
1.1 |
GO:0021902 |
commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 |
0.2 |
GO:0008612 |
peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.1 |
0.3 |
GO:0070127 |
tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.1 |
0.5 |
GO:0016081 |
synaptic vesicle docking(GO:0016081) |
| 0.1 |
0.2 |
GO:0090071 |
negative regulation of ribosome biogenesis(GO:0090071) |
| 0.1 |
0.5 |
GO:0032596 |
protein transport into membrane raft(GO:0032596) dsRNA transport(GO:0033227) |
| 0.1 |
0.3 |
GO:1902683 |
regulation of receptor localization to synapse(GO:1902683) |
| 0.1 |
0.2 |
GO:0044849 |
estrous cycle(GO:0044849) |
| 0.1 |
0.1 |
GO:0035880 |
embryonic nail plate morphogenesis(GO:0035880) |
| 0.1 |
0.2 |
GO:0045659 |
regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.1 |
0.2 |
GO:0032066 |
nucleolus to nucleoplasm transport(GO:0032066) |
| 0.1 |
0.3 |
GO:0032346 |
positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) |
| 0.1 |
0.2 |
GO:0010989 |
negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.1 |
0.3 |
GO:0019086 |
late viral transcription(GO:0019086) |
| 0.1 |
0.3 |
GO:2001204 |
regulation of osteoclast development(GO:2001204) |
| 0.1 |
0.3 |
GO:1902861 |
copper ion import into cell(GO:1902861) |
| 0.1 |
0.2 |
GO:0035696 |
monocyte extravasation(GO:0035696) regulation of monocyte extravasation(GO:2000437) |
| 0.1 |
0.7 |
GO:0000185 |
activation of MAPKKK activity(GO:0000185) |
| 0.0 |
0.3 |
GO:0070166 |
enamel mineralization(GO:0070166) |
| 0.0 |
0.1 |
GO:0046122 |
purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.0 |
0.2 |
GO:1901069 |
guanosine-containing compound catabolic process(GO:1901069) |
| 0.0 |
0.5 |
GO:0019511 |
peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 |
0.5 |
GO:0032096 |
negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.0 |
0.2 |
GO:2000574 |
regulation of microtubule motor activity(GO:2000574) |
| 0.0 |
0.1 |
GO:0001732 |
formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 |
0.2 |
GO:0006049 |
UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 |
0.3 |
GO:2000002 |
negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.0 |
0.3 |
GO:0006102 |
isocitrate metabolic process(GO:0006102) |
| 0.0 |
0.2 |
GO:0060715 |
syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.0 |
0.1 |
GO:0019043 |
positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) establishment of viral latency(GO:0019043) |
| 0.0 |
0.2 |
GO:0051045 |
negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 |
0.3 |
GO:0060267 |
positive regulation of respiratory burst(GO:0060267) |
| 0.0 |
0.2 |
GO:1901837 |
negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.0 |
0.2 |
GO:0090309 |
positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 |
0.3 |
GO:0031665 |
negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 |
0.2 |
GO:0006933 |
negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 |
0.2 |
GO:2000672 |
negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 |
0.2 |
GO:0036438 |
maintenance of lens transparency(GO:0036438) |
| 0.0 |
0.1 |
GO:0002578 |
negative regulation of antigen processing and presentation(GO:0002578) |
| 0.0 |
0.2 |
GO:0070574 |
cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 |
0.1 |
GO:0007521 |
muscle cell fate determination(GO:0007521) cellular response to parathyroid hormone stimulus(GO:0071374) positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.0 |
0.4 |
GO:0048680 |
neuron remodeling(GO:0016322) positive regulation of axon regeneration(GO:0048680) |
| 0.0 |
0.2 |
GO:1901727 |
positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 |
0.1 |
GO:0050859 |
negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.0 |
0.3 |
GO:0006265 |
DNA topological change(GO:0006265) |
| 0.0 |
0.1 |
GO:0070317 |
negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 |
0.1 |
GO:0016561 |
protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.0 |
0.1 |
GO:1900020 |
regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.0 |
0.2 |
GO:0016255 |
attachment of GPI anchor to protein(GO:0016255) |
| 0.0 |
0.1 |
GO:0014735 |
regulation of muscle atrophy(GO:0014735) |
| 0.0 |
0.1 |
GO:1902774 |
late endosome to lysosome transport(GO:1902774) |
| 0.0 |
0.2 |
GO:0006680 |
glucosylceramide catabolic process(GO:0006680) |
| 0.0 |
0.1 |
GO:0001302 |
replicative cell aging(GO:0001302) |
| 0.0 |
0.5 |
GO:0050765 |
negative regulation of phagocytosis(GO:0050765) |
| 0.0 |
0.4 |
GO:0048280 |
vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 |
0.2 |
GO:0031119 |
tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 |
0.1 |
GO:0060075 |
regulation of resting membrane potential(GO:0060075) |
| 0.0 |
0.1 |
GO:1902255 |
positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 0.0 |
0.4 |
GO:0071577 |
zinc II ion transmembrane transport(GO:0071577) |
| 0.0 |
0.3 |
GO:0034497 |
protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 |
0.1 |
GO:0034214 |
protein hexamerization(GO:0034214) |
| 0.0 |
0.0 |
GO:0036324 |
vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.0 |
0.1 |
GO:0051031 |
tRNA transport(GO:0051031) |
| 0.0 |
0.3 |
GO:0061299 |
retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 |
0.2 |
GO:0051601 |
exocyst localization(GO:0051601) |
| 0.0 |
0.2 |
GO:0071712 |
ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 |
0.7 |
GO:0006783 |
heme biosynthetic process(GO:0006783) |
| 0.0 |
0.2 |
GO:0000395 |
mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 |
0.1 |
GO:2001180 |
regulation of interleukin-18 production(GO:0032661) negative regulation of interleukin-18 production(GO:0032701) negative regulation of interleukin-10 secretion(GO:2001180) |
| 0.0 |
0.3 |
GO:0045651 |
positive regulation of macrophage differentiation(GO:0045651) |
| 0.0 |
0.2 |
GO:2000643 |
positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.0 |
0.6 |
GO:0001893 |
maternal placenta development(GO:0001893) |
| 0.0 |
0.2 |
GO:0018344 |
protein geranylgeranylation(GO:0018344) |
| 0.0 |
0.2 |
GO:0051001 |
negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.0 |
0.3 |
GO:0045792 |
negative regulation of cell size(GO:0045792) |
| 0.0 |
0.1 |
GO:0039530 |
MDA-5 signaling pathway(GO:0039530) |
| 0.0 |
0.1 |
GO:0006556 |
S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 |
0.1 |
GO:0051531 |
NFAT protein import into nucleus(GO:0051531) regulation of NFAT protein import into nucleus(GO:0051532) |
| 0.0 |
0.4 |
GO:0060707 |
trophoblast giant cell differentiation(GO:0060707) |
| 0.0 |
0.2 |
GO:0090085 |
regulation of protein deubiquitination(GO:0090085) |
| 0.0 |
0.2 |
GO:0006488 |
dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 |
0.3 |
GO:0030574 |
collagen catabolic process(GO:0030574) |
| 0.0 |
0.1 |
GO:0023035 |
CD40 signaling pathway(GO:0023035) |
| 0.0 |
0.4 |
GO:0030213 |
hyaluronan biosynthetic process(GO:0030213) |
| 0.0 |
0.3 |
GO:1901898 |
negative regulation of relaxation of muscle(GO:1901078) regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 |
0.1 |
GO:0006701 |
progesterone biosynthetic process(GO:0006701) |
| 0.0 |
0.1 |
GO:1903377 |
negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 |
0.2 |
GO:0000479 |
endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.0 |
0.0 |
GO:0034287 |
detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 |
0.1 |
GO:0035845 |
photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 |
0.1 |
GO:2001187 |
positive regulation of CD8-positive, alpha-beta T cell activation(GO:2001187) |
| 0.0 |
0.4 |
GO:0045026 |
plasma membrane fusion(GO:0045026) |
| 0.0 |
0.1 |
GO:0060744 |
thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 |
0.2 |
GO:0021942 |
radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 |
0.2 |
GO:0034472 |
snRNA 3'-end processing(GO:0034472) |
| 0.0 |
0.1 |
GO:1900026 |
positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 |
0.1 |
GO:0048102 |
autophagic cell death(GO:0048102) |
| 0.0 |
0.8 |
GO:0047496 |
vesicle transport along microtubule(GO:0047496) |
| 0.0 |
0.1 |
GO:0007256 |
activation of JNKK activity(GO:0007256) |
| 0.0 |
0.1 |
GO:0090370 |
negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 |
0.2 |
GO:0006662 |
glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.0 |
0.1 |
GO:0089711 |
L-glutamate transmembrane transport(GO:0089711) |
| 0.0 |
0.1 |
GO:1902775 |
mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 |
0.1 |
GO:0090241 |
negative regulation of histone H4 acetylation(GO:0090241) |
| 0.0 |
0.0 |
GO:0001866 |
NK T cell proliferation(GO:0001866) |
| 0.0 |
0.1 |
GO:0010637 |
negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 |
0.1 |
GO:0032811 |
negative regulation of gamma-aminobutyric acid secretion(GO:0014053) regulation of epinephrine secretion(GO:0014060) negative regulation of epinephrine secretion(GO:0032811) |
| 0.0 |
0.1 |
GO:0007016 |
cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 |
0.1 |
GO:0006817 |
phosphate ion transport(GO:0006817) |
| 0.0 |
0.1 |
GO:0051534 |
negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 |
0.1 |
GO:0006776 |
vitamin A metabolic process(GO:0006776) |
| 0.0 |
0.1 |
GO:0010756 |
positive regulation of plasminogen activation(GO:0010756) |
| 0.0 |
0.0 |
GO:1903416 |
response to glycoside(GO:1903416) |
| 0.0 |
0.1 |
GO:0051014 |
actin filament severing(GO:0051014) |
| 0.0 |
0.0 |
GO:2001053 |
regulation of mesenchymal cell apoptotic process(GO:2001053) negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.0 |
0.2 |
GO:0046855 |
phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 |
0.1 |
GO:0035646 |
endosome to melanosome transport(GO:0035646) endosome to pigment granule transport(GO:0043485) pigment granule maturation(GO:0048757) |
| 0.0 |
0.2 |
GO:0030157 |
pancreatic juice secretion(GO:0030157) |
| 0.0 |
0.1 |
GO:0030259 |
lipid glycosylation(GO:0030259) |
| 0.0 |
0.1 |
GO:0006438 |
valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 |
0.1 |
GO:0017183 |
peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 |
0.1 |
GO:1902202 |
regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) positive regulation of receptor catabolic process(GO:2000646) |
| 0.0 |
0.1 |
GO:0090164 |
asymmetric Golgi ribbon formation(GO:0090164) |
| 0.0 |
0.1 |
GO:0009449 |
gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 |
0.1 |
GO:0071225 |
cellular response to muramyl dipeptide(GO:0071225) |
| 0.0 |
0.2 |
GO:0010388 |
protein deneddylation(GO:0000338) cullin deneddylation(GO:0010388) |
| 0.0 |
0.0 |
GO:0042998 |
positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.0 |
0.1 |
GO:0000491 |
small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 |
0.1 |
GO:0072015 |
glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 |
0.2 |
GO:0001522 |
pseudouridine synthesis(GO:0001522) |
| 0.0 |
0.1 |
GO:0033313 |
meiotic cell cycle checkpoint(GO:0033313) |
| 0.0 |
0.1 |
GO:0006013 |
mannose metabolic process(GO:0006013) |
| 0.0 |
0.1 |
GO:0070995 |
NADPH oxidation(GO:0070995) |
| 0.0 |
0.1 |
GO:0007000 |
nucleolus organization(GO:0007000) |
| 0.0 |
0.4 |
GO:0031529 |
ruffle organization(GO:0031529) |
| 0.0 |
0.1 |
GO:1902287 |
semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 |
0.3 |
GO:0001921 |
positive regulation of receptor recycling(GO:0001921) |
| 0.0 |
0.0 |
GO:0098915 |
membrane repolarization during ventricular cardiac muscle cell action potential(GO:0098915) |
| 0.0 |
0.1 |
GO:0048711 |
positive regulation of astrocyte differentiation(GO:0048711) |
| 0.0 |
0.2 |
GO:0000028 |
ribosomal small subunit assembly(GO:0000028) |
| 0.0 |
0.0 |
GO:0002002 |
regulation of angiotensin levels in blood(GO:0002002) |
| 0.0 |
0.3 |
GO:0003333 |
amino acid transmembrane transport(GO:0003333) |
| 0.0 |
0.1 |
GO:1901621 |
negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 |
0.1 |
GO:2000491 |
positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 |
0.0 |
GO:0042255 |
ribosome assembly(GO:0042255) |
| 0.0 |
0.0 |
GO:0045618 |
positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.0 |
0.1 |
GO:0034244 |
negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 |
0.4 |
GO:0048843 |
negative regulation of axon extension involved in axon guidance(GO:0048843) |
| 0.0 |
0.0 |
GO:0018103 |
protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 |
0.0 |
GO:0002309 |
T cell proliferation involved in immune response(GO:0002309) |
| 0.0 |
0.0 |
GO:0010626 |
negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.0 |
0.1 |
GO:0000301 |
retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 |
0.1 |
GO:0043302 |
positive regulation of leukocyte degranulation(GO:0043302) |
| 0.0 |
0.1 |
GO:0046929 |
negative regulation of neurotransmitter secretion(GO:0046929) |