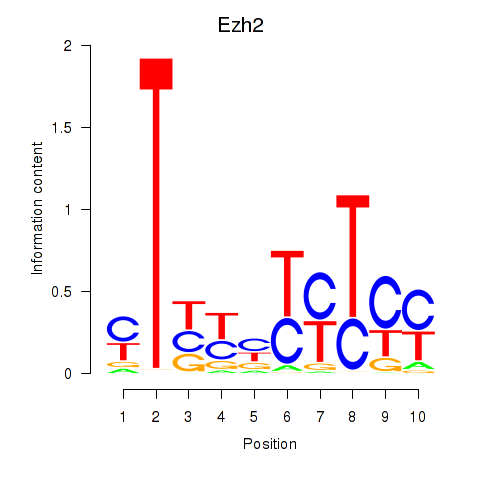
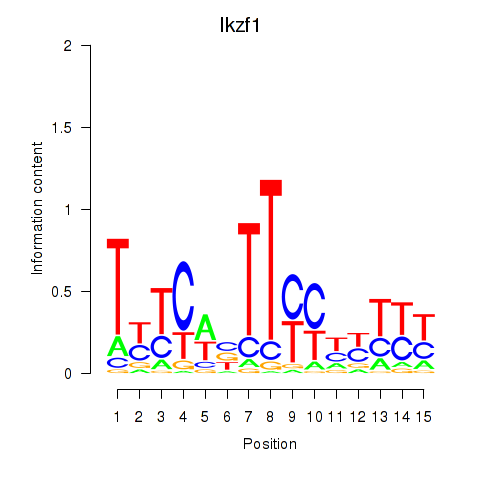
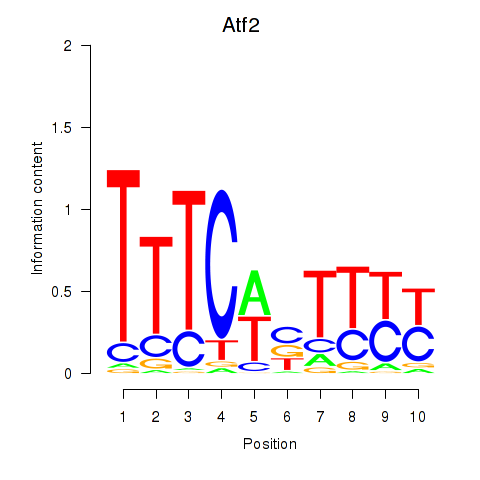
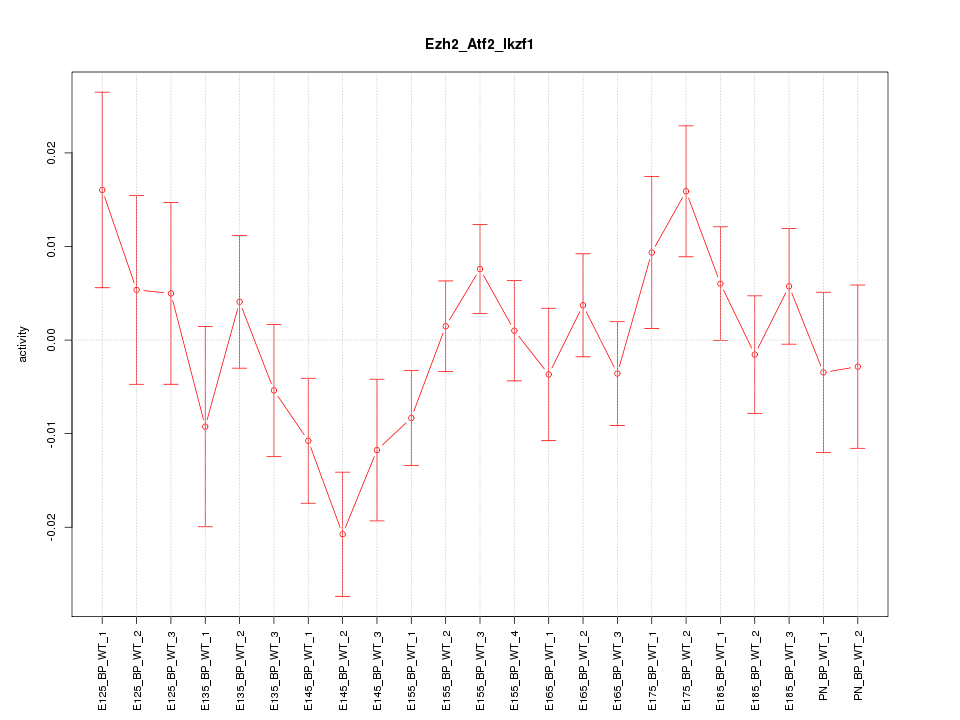
| 4.6 |
13.7 |
GO:0090425 |
hepatocyte cell migration(GO:0002194) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 1.2 |
3.7 |
GO:0060067 |
cervix development(GO:0060067) |
| 1.0 |
3.0 |
GO:0007210 |
serotonin receptor signaling pathway(GO:0007210) |
| 0.5 |
2.5 |
GO:0002457 |
T cell antigen processing and presentation(GO:0002457) |
| 0.4 |
2.2 |
GO:0003404 |
optic vesicle morphogenesis(GO:0003404) |
| 0.4 |
2.1 |
GO:0097116 |
gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.3 |
1.0 |
GO:0007525 |
somatic muscle development(GO:0007525) |
| 0.3 |
1.2 |
GO:0021698 |
cerebellar cortex structural organization(GO:0021698) |
| 0.3 |
4.1 |
GO:0021979 |
hypothalamus cell differentiation(GO:0021979) |
| 0.3 |
1.1 |
GO:0055099 |
regulation of Cdc42 protein signal transduction(GO:0032489) response to high density lipoprotein particle(GO:0055099) platelet dense granule organization(GO:0060155) |
| 0.3 |
0.8 |
GO:1903225 |
negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.3 |
1.1 |
GO:0097393 |
post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 0.3 |
9.9 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.3 |
1.1 |
GO:0032512 |
regulation of protein phosphatase type 2B activity(GO:0032512) positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.3 |
1.0 |
GO:1904425 |
negative regulation of GTP binding(GO:1904425) |
| 0.3 |
1.3 |
GO:0021965 |
spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.2 |
0.7 |
GO:0060244 |
negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.2 |
1.0 |
GO:0007158 |
neuron cell-cell adhesion(GO:0007158) |
| 0.2 |
0.7 |
GO:0008228 |
opsonization(GO:0008228) |
| 0.2 |
0.7 |
GO:2000686 |
regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.2 |
1.4 |
GO:0097105 |
presynaptic membrane assembly(GO:0097105) |
| 0.2 |
2.3 |
GO:0015721 |
bile acid and bile salt transport(GO:0015721) |
| 0.2 |
5.5 |
GO:0010172 |
embryonic body morphogenesis(GO:0010172) |
| 0.2 |
0.2 |
GO:0070315 |
G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.2 |
0.7 |
GO:0002331 |
pre-B cell allelic exclusion(GO:0002331) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.2 |
0.9 |
GO:1903984 |
positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.2 |
0.7 |
GO:1900736 |
regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900736) |
| 0.2 |
1.2 |
GO:2000790 |
regulation of mesenchymal cell proliferation involved in lung development(GO:2000790) negative regulation of mesenchymal cell proliferation involved in lung development(GO:2000791) |
| 0.2 |
0.8 |
GO:1903251 |
multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.2 |
1.2 |
GO:0071883 |
activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.2 |
0.8 |
GO:0046552 |
eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.2 |
0.5 |
GO:1902528 |
regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.2 |
0.5 |
GO:0003221 |
right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.2 |
2.8 |
GO:0048387 |
negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.2 |
1.3 |
GO:0048625 |
myoblast fate commitment(GO:0048625) |
| 0.2 |
0.8 |
GO:0090310 |
negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.2 |
1.3 |
GO:0071557 |
histone H3-K27 demethylation(GO:0071557) |
| 0.2 |
0.8 |
GO:0090188 |
negative regulation of pancreatic juice secretion(GO:0090188) |
| 0.2 |
0.3 |
GO:0071360 |
cellular response to exogenous dsRNA(GO:0071360) |
| 0.1 |
0.9 |
GO:0019336 |
phenol-containing compound catabolic process(GO:0019336) |
| 0.1 |
0.4 |
GO:1900365 |
positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.1 |
0.3 |
GO:1903243 |
negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) |
| 0.1 |
0.6 |
GO:1903378 |
positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.1 |
0.4 |
GO:0019085 |
early viral transcription(GO:0019085) |
| 0.1 |
0.1 |
GO:0070094 |
positive regulation of glucagon secretion(GO:0070094) |
| 0.1 |
0.4 |
GO:0044849 |
estrous cycle(GO:0044849) |
| 0.1 |
0.3 |
GO:1902336 |
neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) positive regulation of retinal ganglion cell axon guidance(GO:1902336) |
| 0.1 |
0.4 |
GO:0030208 |
dermatan sulfate biosynthetic process(GO:0030208) |
| 0.1 |
0.4 |
GO:0030167 |
proteoglycan catabolic process(GO:0030167) |
| 0.1 |
2.0 |
GO:0021796 |
cerebral cortex regionalization(GO:0021796) |
| 0.1 |
0.5 |
GO:0051088 |
PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.1 |
0.7 |
GO:0043415 |
positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.1 |
0.5 |
GO:0060282 |
positive regulation of oocyte development(GO:0060282) |
| 0.1 |
0.4 |
GO:0051464 |
negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) positive regulation of cortisol secretion(GO:0051464) |
| 0.1 |
0.5 |
GO:0060753 |
regulation of mast cell chemotaxis(GO:0060753) |
| 0.1 |
0.7 |
GO:0051694 |
pointed-end actin filament capping(GO:0051694) |
| 0.1 |
0.3 |
GO:0048014 |
Tie signaling pathway(GO:0048014) |
| 0.1 |
0.3 |
GO:0050703 |
interleukin-1 alpha secretion(GO:0050703) |
| 0.1 |
0.2 |
GO:0060741 |
prostate gland stromal morphogenesis(GO:0060741) |
| 0.1 |
0.3 |
GO:0048859 |
smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) formation of anatomical boundary(GO:0048859) positive regulation of epithelial cell proliferation involved in prostate gland development(GO:0060769) negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.1 |
0.5 |
GO:0046684 |
response to pyrethroid(GO:0046684) |
| 0.1 |
0.4 |
GO:0015793 |
glycerol transport(GO:0015793) |
| 0.1 |
0.2 |
GO:0032488 |
Cdc42 protein signal transduction(GO:0032488) |
| 0.1 |
0.8 |
GO:0006620 |
posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 |
0.3 |
GO:0015712 |
hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 |
0.3 |
GO:0015910 |
peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.1 |
0.2 |
GO:0001543 |
ovarian follicle rupture(GO:0001543) |
| 0.1 |
1.5 |
GO:0042759 |
long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 |
0.2 |
GO:0002578 |
negative regulation of antigen processing and presentation(GO:0002578) |
| 0.1 |
0.7 |
GO:0030718 |
germ-line stem cell population maintenance(GO:0030718) |
| 0.1 |
4.0 |
GO:0006376 |
mRNA splice site selection(GO:0006376) |
| 0.1 |
0.7 |
GO:0030579 |
ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 |
0.4 |
GO:0055071 |
cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.1 |
0.2 |
GO:0008065 |
establishment of blood-nerve barrier(GO:0008065) |
| 0.1 |
0.4 |
GO:2000809 |
positive regulation of presynaptic membrane organization(GO:1901631) positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.1 |
0.4 |
GO:0033136 |
serine phosphorylation of STAT3 protein(GO:0033136) |
| 0.1 |
0.3 |
GO:0008594 |
photoreceptor cell morphogenesis(GO:0008594) |
| 0.1 |
0.3 |
GO:0050904 |
diapedesis(GO:0050904) |
| 0.1 |
0.4 |
GO:2001168 |
regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.1 |
0.4 |
GO:0060762 |
regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.1 |
0.2 |
GO:0002730 |
regulation of dendritic cell cytokine production(GO:0002730) |
| 0.1 |
1.2 |
GO:0002089 |
lens morphogenesis in camera-type eye(GO:0002089) |
| 0.1 |
0.3 |
GO:0060083 |
smooth muscle contraction involved in micturition(GO:0060083) |
| 0.1 |
1.4 |
GO:0032011 |
ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 |
0.2 |
GO:0021837 |
motogenic signaling involved in postnatal olfactory bulb interneuron migration(GO:0021837) positive regulation of mitotic cell cycle DNA replication(GO:1903465) |
| 0.1 |
0.5 |
GO:0007342 |
fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.1 |
0.5 |
GO:0071699 |
olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.1 |
0.2 |
GO:0021691 |
cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.1 |
0.6 |
GO:0032876 |
negative regulation of DNA endoreduplication(GO:0032876) |
| 0.1 |
0.7 |
GO:0071420 |
cellular response to histamine(GO:0071420) |
| 0.1 |
0.3 |
GO:0034454 |
microtubule anchoring at centrosome(GO:0034454) |
| 0.1 |
0.3 |
GO:1903800 |
astrocyte activation(GO:0048143) positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 |
0.6 |
GO:0019227 |
neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 |
0.1 |
GO:0032836 |
glomerular basement membrane development(GO:0032836) |
| 0.1 |
0.2 |
GO:1902177 |
positive regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902177) |
| 0.1 |
0.1 |
GO:0048686 |
regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.1 |
0.6 |
GO:0018344 |
protein geranylgeranylation(GO:0018344) |
| 0.1 |
0.4 |
GO:0090074 |
negative regulation of protein homodimerization activity(GO:0090074) |
| 0.1 |
0.3 |
GO:0035063 |
nuclear speck organization(GO:0035063) |
| 0.1 |
0.1 |
GO:0014016 |
neuroblast differentiation(GO:0014016) |
| 0.1 |
0.3 |
GO:0035616 |
histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.1 |
0.6 |
GO:0031339 |
negative regulation of vesicle fusion(GO:0031339) |
| 0.1 |
0.3 |
GO:0061073 |
ciliary body morphogenesis(GO:0061073) |
| 0.1 |
0.3 |
GO:0046501 |
protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.1 |
0.2 |
GO:0016052 |
carbohydrate catabolic process(GO:0016052) |
| 0.1 |
1.2 |
GO:0019835 |
cytolysis(GO:0019835) |
| 0.1 |
6.2 |
GO:0032092 |
positive regulation of protein binding(GO:0032092) |
| 0.1 |
0.5 |
GO:0061368 |
behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.1 |
0.3 |
GO:0098735 |
positive regulation of the force of heart contraction(GO:0098735) |
| 0.1 |
0.4 |
GO:0003406 |
retinal pigment epithelium development(GO:0003406) |
| 0.1 |
0.2 |
GO:2000858 |
mineralocorticoid secretion(GO:0035931) aldosterone secretion(GO:0035932) regulation of mineralocorticoid secretion(GO:2000855) positive regulation of mineralocorticoid secretion(GO:2000857) regulation of aldosterone secretion(GO:2000858) positive regulation of aldosterone secretion(GO:2000860) |
| 0.1 |
0.2 |
GO:0032790 |
ribosome disassembly(GO:0032790) |
| 0.1 |
2.4 |
GO:0090162 |
establishment of epithelial cell polarity(GO:0090162) |
| 0.1 |
0.2 |
GO:0009957 |
epidermal cell fate specification(GO:0009957) |
| 0.1 |
1.3 |
GO:0035641 |
locomotory exploration behavior(GO:0035641) |
| 0.1 |
0.1 |
GO:0050883 |
musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.1 |
0.4 |
GO:0016584 |
nucleosome positioning(GO:0016584) |
| 0.1 |
0.2 |
GO:0014719 |
skeletal muscle satellite cell activation(GO:0014719) |
| 0.1 |
0.2 |
GO:0033326 |
cerebrospinal fluid secretion(GO:0033326) |
| 0.1 |
0.3 |
GO:0045654 |
positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.1 |
0.2 |
GO:0045162 |
clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 |
0.2 |
GO:0090258 |
negative regulation of mitochondrial fission(GO:0090258) |
| 0.1 |
0.3 |
GO:0001542 |
ovulation from ovarian follicle(GO:0001542) |
| 0.1 |
0.2 |
GO:0048633 |
positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.1 |
0.3 |
GO:0045743 |
positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.1 |
0.3 |
GO:0048934 |
peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 |
0.2 |
GO:0019858 |
cytosine metabolic process(GO:0019858) |
| 0.1 |
0.3 |
GO:0070072 |
vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 |
0.2 |
GO:1903288 |
regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) |
| 0.1 |
0.2 |
GO:0030321 |
transepithelial chloride transport(GO:0030321) |
| 0.0 |
0.2 |
GO:2000325 |
regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.0 |
0.2 |
GO:2001045 |
negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 |
0.0 |
GO:2000574 |
regulation of microtubule motor activity(GO:2000574) |
| 0.0 |
0.3 |
GO:0010528 |
regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.0 |
0.4 |
GO:0070102 |
interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.0 |
0.4 |
GO:0002674 |
negative regulation of acute inflammatory response(GO:0002674) |
| 0.0 |
0.2 |
GO:0097167 |
circadian regulation of translation(GO:0097167) |
| 0.0 |
0.2 |
GO:0051890 |
regulation of cardioblast differentiation(GO:0051890) |
| 0.0 |
0.5 |
GO:0001672 |
regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 |
0.5 |
GO:0090129 |
positive regulation of synapse maturation(GO:0090129) |
| 0.0 |
0.6 |
GO:0060049 |
regulation of protein glycosylation(GO:0060049) |
| 0.0 |
0.3 |
GO:1903887 |
motile primary cilium assembly(GO:1903887) |
| 0.0 |
0.2 |
GO:0090399 |
replicative senescence(GO:0090399) |
| 0.0 |
0.6 |
GO:0009312 |
oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 |
0.3 |
GO:1901844 |
regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.0 |
0.2 |
GO:0015867 |
ATP transport(GO:0015867) |
| 0.0 |
0.2 |
GO:0070213 |
protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 |
3.4 |
GO:0001578 |
microtubule bundle formation(GO:0001578) |
| 0.0 |
0.5 |
GO:0046697 |
decidualization(GO:0046697) |
| 0.0 |
0.2 |
GO:1901475 |
pyruvate transmembrane transport(GO:1901475) |
| 0.0 |
0.2 |
GO:0060073 |
regulation of the force of heart contraction by chemical signal(GO:0003057) micturition(GO:0060073) |
| 0.0 |
0.2 |
GO:0042447 |
hormone catabolic process(GO:0042447) |
| 0.0 |
0.2 |
GO:1903898 |
positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.0 |
0.2 |
GO:0003365 |
establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.0 |
0.3 |
GO:0048149 |
behavioral response to ethanol(GO:0048149) |
| 0.0 |
0.1 |
GO:0050925 |
negative regulation of negative chemotaxis(GO:0050925) |
| 0.0 |
0.5 |
GO:0048520 |
positive regulation of behavior(GO:0048520) |
| 0.0 |
0.4 |
GO:0007175 |
negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 |
0.1 |
GO:0010820 |
positive regulation of T cell chemotaxis(GO:0010820) |
| 0.0 |
0.1 |
GO:0033030 |
negative regulation of neutrophil apoptotic process(GO:0033030) |
| 0.0 |
0.4 |
GO:0061470 |
T follicular helper cell differentiation(GO:0061470) |
| 0.0 |
0.2 |
GO:1901341 |
activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 |
0.2 |
GO:0060718 |
chorionic trophoblast cell differentiation(GO:0060718) |
| 0.0 |
1.7 |
GO:0007628 |
adult walking behavior(GO:0007628) walking behavior(GO:0090659) |
| 0.0 |
0.5 |
GO:2001224 |
positive regulation of neuron migration(GO:2001224) |
| 0.0 |
0.2 |
GO:0090527 |
actin filament reorganization(GO:0090527) |
| 0.0 |
0.3 |
GO:0035520 |
monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 |
0.1 |
GO:0071442 |
positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 |
0.2 |
GO:0060903 |
positive regulation of meiosis I(GO:0060903) |
| 0.0 |
0.1 |
GO:0060075 |
regulation of resting membrane potential(GO:0060075) |
| 0.0 |
0.1 |
GO:0045059 |
positive thymic T cell selection(GO:0045059) |
| 0.0 |
0.3 |
GO:0000083 |
regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 |
1.2 |
GO:0000186 |
activation of MAPKK activity(GO:0000186) |
| 0.0 |
0.3 |
GO:0014841 |
skeletal muscle satellite cell proliferation(GO:0014841) |
| 0.0 |
0.1 |
GO:2001181 |
positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.0 |
0.2 |
GO:0061086 |
negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 |
0.2 |
GO:0031122 |
cytoplasmic microtubule organization(GO:0031122) |
| 0.0 |
0.2 |
GO:1904754 |
positive regulation of vascular associated smooth muscle cell migration(GO:1904754) |
| 0.0 |
0.2 |
GO:0007258 |
JUN phosphorylation(GO:0007258) |
| 0.0 |
0.3 |
GO:1903608 |
protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 |
0.2 |
GO:0007016 |
cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 |
0.2 |
GO:0090383 |
phagosome acidification(GO:0090383) |
| 0.0 |
0.2 |
GO:0021780 |
oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 |
0.1 |
GO:0097709 |
connective tissue replacement involved in inflammatory response wound healing(GO:0002248) connective tissue replacement(GO:0097709) |
| 0.0 |
0.1 |
GO:0007171 |
activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 |
0.1 |
GO:0090370 |
negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 |
1.7 |
GO:0072661 |
protein targeting to plasma membrane(GO:0072661) |
| 0.0 |
0.2 |
GO:0097368 |
establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 |
1.0 |
GO:0097120 |
receptor localization to synapse(GO:0097120) |
| 0.0 |
0.2 |
GO:0031508 |
pericentric heterochromatin assembly(GO:0031508) |
| 0.0 |
0.5 |
GO:0001731 |
formation of translation preinitiation complex(GO:0001731) |
| 0.0 |
0.1 |
GO:0048162 |
preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.0 |
0.1 |
GO:1902950 |
regulation of dendritic spine maintenance(GO:1902950) positive regulation of dendritic spine maintenance(GO:1902952) |
| 0.0 |
2.5 |
GO:0043488 |
regulation of mRNA stability(GO:0043488) |
| 0.0 |
0.0 |
GO:2000138 |
positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.0 |
0.1 |
GO:0031547 |
brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.0 |
0.5 |
GO:0090036 |
regulation of protein kinase C signaling(GO:0090036) |
| 0.0 |
0.3 |
GO:2000480 |
negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 |
0.0 |
GO:0000821 |
regulation of arginine metabolic process(GO:0000821) |
| 0.0 |
0.5 |
GO:0070816 |
phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 |
0.1 |
GO:0090091 |
positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 |
0.4 |
GO:0048642 |
negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.0 |
0.1 |
GO:0010808 |
positive regulation of synaptic vesicle priming(GO:0010808) positive regulation of synaptic vesicle exocytosis(GO:2000302) |
| 0.0 |
0.2 |
GO:0006686 |
sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 |
0.6 |
GO:0001886 |
endothelial cell morphogenesis(GO:0001886) |
| 0.0 |
0.2 |
GO:0007296 |
vitellogenesis(GO:0007296) |
| 0.0 |
0.4 |
GO:0071378 |
growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 |
0.9 |
GO:0003333 |
amino acid transmembrane transport(GO:0003333) |
| 0.0 |
0.1 |
GO:0051013 |
microtubule severing(GO:0051013) |
| 0.0 |
0.3 |
GO:0035563 |
positive regulation of chromatin binding(GO:0035563) |
| 0.0 |
0.3 |
GO:0090050 |
positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 |
0.5 |
GO:0050829 |
defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 |
0.1 |
GO:0030259 |
lipid glycosylation(GO:0030259) |
| 0.0 |
0.3 |
GO:0060009 |
Sertoli cell development(GO:0060009) |
| 0.0 |
0.1 |
GO:0016339 |
calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 |
0.4 |
GO:0006828 |
manganese ion transport(GO:0006828) |
| 0.0 |
0.1 |
GO:0097091 |
synaptic vesicle clustering(GO:0097091) |
| 0.0 |
0.4 |
GO:0001504 |
neurotransmitter uptake(GO:0001504) |
| 0.0 |
0.1 |
GO:0070829 |
heterochromatin maintenance(GO:0070829) |
| 0.0 |
0.5 |
GO:0016486 |
peptide hormone processing(GO:0016486) |
| 0.0 |
0.6 |
GO:0032292 |
myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 |
0.3 |
GO:0043374 |
CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 |
0.2 |
GO:0048733 |
sebaceous gland development(GO:0048733) |
| 0.0 |
0.6 |
GO:0007588 |
excretion(GO:0007588) |
| 0.0 |
0.6 |
GO:0010842 |
retina layer formation(GO:0010842) |
| 0.0 |
0.3 |
GO:0006273 |
lagging strand elongation(GO:0006273) |
| 0.0 |
0.5 |
GO:0048305 |
immunoglobulin secretion(GO:0048305) |
| 0.0 |
0.0 |
GO:0060018 |
astrocyte fate commitment(GO:0060018) |
| 0.0 |
1.1 |
GO:0010811 |
positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 |
0.2 |
GO:0042590 |
antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.0 |
0.2 |
GO:0021785 |
branchiomotor neuron axon guidance(GO:0021785) |
| 0.0 |
0.2 |
GO:1901162 |
serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 |
0.2 |
GO:1990440 |
positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 |
0.2 |
GO:0046602 |
regulation of mitotic centrosome separation(GO:0046602) |
| 0.0 |
0.1 |
GO:0032463 |
negative regulation of protein homooligomerization(GO:0032463) |
| 0.0 |
0.1 |
GO:0006664 |
glycolipid metabolic process(GO:0006664) liposaccharide metabolic process(GO:1903509) |
| 0.0 |
0.2 |
GO:0035188 |
blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 |
0.5 |
GO:0071108 |
protein K48-linked deubiquitination(GO:0071108) |
| 0.0 |
0.4 |
GO:0032148 |
activation of protein kinase B activity(GO:0032148) |
| 0.0 |
0.1 |
GO:0045742 |
positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) positive regulation of ERBB signaling pathway(GO:1901186) |
| 0.0 |
0.2 |
GO:0009404 |
toxin metabolic process(GO:0009404) |
| 0.0 |
0.5 |
GO:0032801 |
receptor catabolic process(GO:0032801) |
| 0.0 |
0.1 |
GO:0010593 |
negative regulation of lamellipodium assembly(GO:0010593) |
| 0.0 |
0.3 |
GO:0070914 |
UV-damage excision repair(GO:0070914) |
| 0.0 |
0.2 |
GO:0043586 |
tongue development(GO:0043586) |
| 0.0 |
2.0 |
GO:0045727 |
positive regulation of translation(GO:0045727) |
| 0.0 |
0.2 |
GO:0018026 |
peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 |
0.7 |
GO:0048741 |
skeletal muscle fiber development(GO:0048741) |
| 0.0 |
0.1 |
GO:0043928 |
exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 |
0.1 |
GO:1904426 |
positive regulation of GTP binding(GO:1904426) regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 0.0 |
0.1 |
GO:0006436 |
tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 |
0.1 |
GO:0032000 |
positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 |
0.2 |
GO:0051482 |
positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 |
0.1 |
GO:0045085 |
negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 |
0.2 |
GO:0006020 |
inositol metabolic process(GO:0006020) |
| 0.0 |
0.2 |
GO:0043084 |
penile erection(GO:0043084) |
| 0.0 |
0.0 |
GO:0002677 |
negative regulation of chronic inflammatory response(GO:0002677) |
| 0.0 |
0.0 |
GO:0003431 |
growth plate cartilage chondrocyte development(GO:0003431) |
| 0.0 |
0.2 |
GO:0033353 |
S-adenosylmethionine cycle(GO:0033353) |
| 0.0 |
0.2 |
GO:0097067 |
cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 |
0.2 |
GO:0032211 |
negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.0 |
0.2 |
GO:0043517 |
positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 |
0.1 |
GO:0090292 |
nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 |
0.3 |
GO:0046827 |
positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 |
0.1 |
GO:1902260 |
negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 |
0.0 |
GO:0060594 |
mammary gland specification(GO:0060594) |
| 0.0 |
0.2 |
GO:0006883 |
cellular sodium ion homeostasis(GO:0006883) |
| 0.0 |
0.2 |
GO:0021869 |
forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 |
0.2 |
GO:0034453 |
microtubule anchoring(GO:0034453) |
| 0.0 |
0.3 |
GO:0042088 |
T-helper 1 type immune response(GO:0042088) |
| 0.0 |
0.1 |
GO:0060330 |
regulation of response to interferon-gamma(GO:0060330) |
| 0.0 |
0.2 |
GO:0090261 |
positive regulation of inclusion body assembly(GO:0090261) |
| 0.0 |
0.1 |
GO:0042517 |
positive regulation of tyrosine phosphorylation of Stat3 protein(GO:0042517) |
| 0.0 |
0.0 |
GO:0060385 |
axonogenesis involved in innervation(GO:0060385) |
| 0.0 |
0.6 |
GO:0006270 |
DNA replication initiation(GO:0006270) |
| 0.0 |
0.1 |
GO:1990928 |
response to amino acid starvation(GO:1990928) |
| 0.0 |
0.2 |
GO:0046545 |
development of primary female sexual characteristics(GO:0046545) |
| 0.0 |
0.1 |
GO:0034433 |
steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.0 |
0.2 |
GO:0071364 |
cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 |
0.1 |
GO:1903301 |
positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 |
0.0 |
GO:0036292 |
DNA rewinding(GO:0036292) |
| 0.0 |
0.1 |
GO:0050667 |
homocysteine metabolic process(GO:0050667) |
| 0.0 |
0.1 |
GO:0046477 |
glycosylceramide catabolic process(GO:0046477) |
| 0.0 |
0.3 |
GO:0008090 |
retrograde axonal transport(GO:0008090) |
| 0.0 |
0.2 |
GO:0035855 |
megakaryocyte development(GO:0035855) |
| 0.0 |
0.1 |
GO:0006547 |
histidine metabolic process(GO:0006547) imidazole-containing compound metabolic process(GO:0052803) |
| 0.0 |
0.1 |
GO:0032486 |
Rap protein signal transduction(GO:0032486) |
| 0.0 |
0.1 |
GO:2000418 |
positive regulation of eosinophil migration(GO:2000418) |
| 0.0 |
0.1 |
GO:1902414 |
protein localization to cell junction(GO:1902414) |
| 0.0 |
0.1 |
GO:0015074 |
DNA integration(GO:0015074) |
| 0.0 |
0.0 |
GO:0033313 |
meiotic cell cycle checkpoint(GO:0033313) |
| 0.0 |
0.4 |
GO:0016180 |
snRNA processing(GO:0016180) |
| 0.0 |
0.2 |
GO:0016973 |
poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 |
0.2 |
GO:0002495 |
antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.0 |
0.3 |
GO:0048747 |
muscle fiber development(GO:0048747) |
| 0.0 |
0.5 |
GO:0000038 |
very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 |
0.0 |
GO:0016598 |
protein arginylation(GO:0016598) |
| 0.0 |
0.0 |
GO:0044333 |
Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.0 |
0.0 |
GO:0022028 |
tangential migration from the subventricular zone to the olfactory bulb(GO:0022028) |
| 0.0 |
0.0 |
GO:0003360 |
brainstem development(GO:0003360) |
| 0.0 |
0.5 |
GO:0030835 |
negative regulation of actin filament depolymerization(GO:0030835) |
| 0.0 |
0.1 |
GO:1902031 |
regulation of NADP metabolic process(GO:1902031) |
| 0.0 |
0.2 |
GO:1903393 |
positive regulation of adherens junction organization(GO:1903393) |
| 0.0 |
0.0 |
GO:1903298 |
regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903297) negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903298) |
| 0.0 |
0.0 |
GO:0097688 |
AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.0 |
0.2 |
GO:0060716 |
labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 |
0.2 |
GO:0007097 |
nuclear migration(GO:0007097) |
| 0.0 |
0.8 |
GO:0006367 |
transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 |
0.1 |
GO:0031297 |
replication fork processing(GO:0031297) |
| 0.0 |
0.2 |
GO:0008209 |
androgen metabolic process(GO:0008209) |
| 0.0 |
0.1 |
GO:0000479 |
endonucleolytic cleavage involved in rRNA processing(GO:0000478) endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.0 |
0.7 |
GO:0060113 |
inner ear receptor cell differentiation(GO:0060113) |
| 0.0 |
0.4 |
GO:0001947 |
heart looping(GO:0001947) |
| 0.0 |
0.0 |
GO:0005513 |
detection of calcium ion(GO:0005513) |
| 0.0 |
0.2 |
GO:2000114 |
regulation of establishment of cell polarity(GO:2000114) |
| 0.0 |
0.2 |
GO:0015693 |
magnesium ion transport(GO:0015693) |
| 0.0 |
0.2 |
GO:0071392 |
cellular response to estradiol stimulus(GO:0071392) |
| 0.0 |
0.1 |
GO:0042636 |
negative regulation of hair cycle(GO:0042636) negative regulation of hair follicle development(GO:0051799) |
| 0.0 |
0.0 |
GO:0001580 |
detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 |
0.3 |
GO:0045116 |
protein neddylation(GO:0045116) |
| 0.0 |
0.1 |
GO:0046085 |
adenosine metabolic process(GO:0046085) |
| 0.0 |
0.2 |
GO:0032528 |
microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.0 |
0.0 |
GO:0036112 |
medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.0 |
0.1 |
GO:1903715 |
regulation of aerobic respiration(GO:1903715) |
| 0.0 |
0.0 |
GO:1901386 |
negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.0 |
0.3 |
GO:0097320 |
membrane tubulation(GO:0097320) |
| 0.0 |
0.2 |
GO:0099500 |
synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 |
0.1 |
GO:0030813 |
positive regulation of nucleotide catabolic process(GO:0030813) positive regulation of glycolytic process(GO:0045821) positive regulation of cofactor metabolic process(GO:0051194) positive regulation of coenzyme metabolic process(GO:0051197) |
| 0.0 |
0.3 |
GO:1900026 |
positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 |
0.2 |
GO:2000001 |
regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 |
0.1 |
GO:0015838 |
amino-acid betaine transport(GO:0015838) |
| 0.0 |
0.1 |
GO:0031055 |
chromatin remodeling at centromere(GO:0031055) |
| 0.0 |
1.0 |
GO:0000725 |
double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 0.0 |
0.1 |
GO:0044091 |
membrane biogenesis(GO:0044091) |
| 0.0 |
0.0 |
GO:0060923 |
cardiac muscle cell fate commitment(GO:0060923) |
| 0.0 |
0.1 |
GO:0097341 |
inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 |
0.1 |
GO:0007214 |
gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 |
0.0 |
GO:1904417 |
regulation of xenophagy(GO:1904415) positive regulation of xenophagy(GO:1904417) |
| 0.0 |
0.1 |
GO:0046601 |
positive regulation of centriole replication(GO:0046601) |
| 0.0 |
0.1 |
GO:0043383 |
negative T cell selection(GO:0043383) negative thymic T cell selection(GO:0045060) |
| 0.0 |
0.0 |
GO:0010801 |
negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 |
0.0 |
GO:0000087 |
mitotic M phase(GO:0000087) mitotic cell cycle phase(GO:0098763) |
| 0.0 |
0.3 |
GO:0033120 |
positive regulation of RNA splicing(GO:0033120) |
| 0.0 |
0.1 |
GO:0042035 |
regulation of cytokine biosynthetic process(GO:0042035) |
| 0.0 |
0.1 |
GO:0051764 |
actin crosslink formation(GO:0051764) |
| 0.0 |
0.0 |
GO:0043553 |
negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 |
0.0 |
GO:1902861 |
copper ion import into cell(GO:1902861) |
| 0.0 |
0.2 |
GO:0030433 |
ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 |
0.1 |
GO:0007064 |
mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 |
0.0 |
GO:0072675 |
osteoclast fusion(GO:0072675) |
| 0.0 |
0.2 |
GO:0008340 |
determination of adult lifespan(GO:0008340) |
| 0.0 |
0.3 |
GO:0070979 |
protein K11-linked ubiquitination(GO:0070979) |
| 0.0 |
0.1 |
GO:0031274 |
regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |