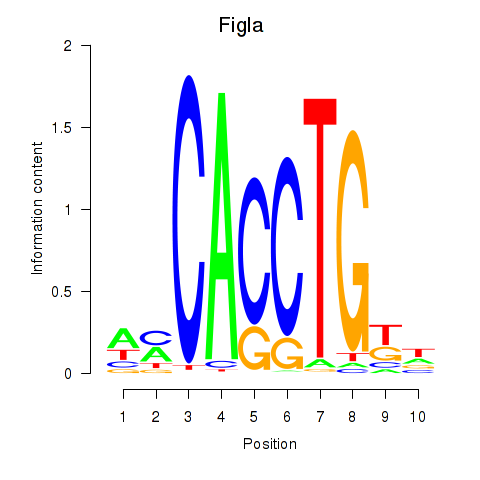
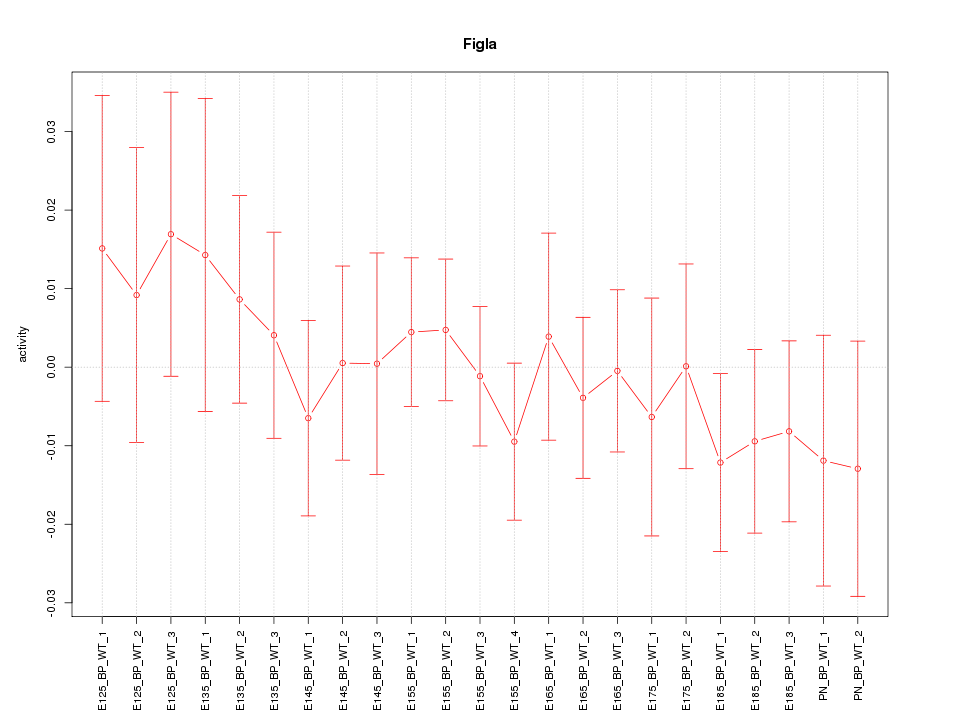
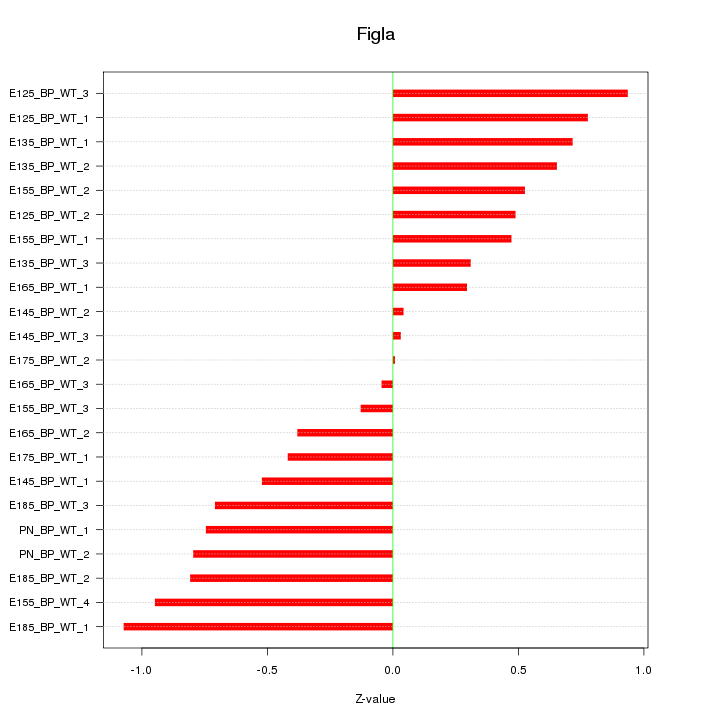
| 0.4 |
2.1 |
GO:0010993 |
regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.4 |
2.0 |
GO:0071205 |
clustering of voltage-gated potassium channels(GO:0045163) protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.4 |
1.5 |
GO:0051311 |
spindle assembly involved in female meiosis(GO:0007056) meiotic metaphase plate congression(GO:0051311) |
| 0.3 |
0.9 |
GO:1904274 |
tricellular tight junction assembly(GO:1904274) |
| 0.3 |
0.9 |
GO:0007181 |
transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.3 |
1.1 |
GO:0043987 |
histone H3-S10 phosphorylation(GO:0043987) |
| 0.3 |
0.8 |
GO:0060023 |
soft palate development(GO:0060023) |
| 0.2 |
1.1 |
GO:0003223 |
ventricular compact myocardium morphogenesis(GO:0003223) proprioception(GO:0019230) |
| 0.2 |
0.8 |
GO:2001271 |
negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.2 |
1.4 |
GO:0000320 |
re-entry into mitotic cell cycle(GO:0000320) |
| 0.2 |
0.6 |
GO:0090258 |
negative regulation of mitochondrial fission(GO:0090258) |
| 0.2 |
0.8 |
GO:1990379 |
lipid transport across blood brain barrier(GO:1990379) |
| 0.2 |
2.0 |
GO:0042487 |
regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.2 |
0.9 |
GO:0045759 |
negative regulation of action potential(GO:0045759) |
| 0.2 |
1.4 |
GO:0005513 |
detection of calcium ion(GO:0005513) |
| 0.2 |
1.0 |
GO:0003383 |
apical constriction(GO:0003383) |
| 0.2 |
1.6 |
GO:0030322 |
stabilization of membrane potential(GO:0030322) |
| 0.2 |
0.6 |
GO:0032439 |
endosome localization(GO:0032439) |
| 0.1 |
0.4 |
GO:0035574 |
histone H4-K20 demethylation(GO:0035574) |
| 0.1 |
1.6 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.1 |
0.4 |
GO:0030167 |
proteoglycan catabolic process(GO:0030167) |
| 0.1 |
0.4 |
GO:0032915 |
positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.1 |
1.0 |
GO:0046826 |
negative regulation of protein export from nucleus(GO:0046826) |
| 0.1 |
0.6 |
GO:0006344 |
maintenance of chromatin silencing(GO:0006344) |
| 0.1 |
0.4 |
GO:0021812 |
neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.1 |
3.4 |
GO:0050873 |
brown fat cell differentiation(GO:0050873) |
| 0.1 |
0.7 |
GO:0090209 |
negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.1 |
0.3 |
GO:0032241 |
positive regulation of nucleobase-containing compound transport(GO:0032241) |
| 0.1 |
1.9 |
GO:0010763 |
positive regulation of fibroblast migration(GO:0010763) |
| 0.1 |
0.6 |
GO:0072017 |
distal tubule development(GO:0072017) |
| 0.1 |
0.4 |
GO:0071361 |
cellular response to ethanol(GO:0071361) |
| 0.1 |
0.5 |
GO:0002315 |
marginal zone B cell differentiation(GO:0002315) |
| 0.1 |
1.3 |
GO:0016339 |
calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 |
0.3 |
GO:0097527 |
necroptotic signaling pathway(GO:0097527) |
| 0.1 |
0.7 |
GO:1902231 |
positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.1 |
2.4 |
GO:0045746 |
negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 |
1.3 |
GO:0032897 |
negative regulation of viral transcription(GO:0032897) |
| 0.1 |
0.4 |
GO:0097491 |
trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.1 |
0.3 |
GO:0007089 |
traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.1 |
0.2 |
GO:0035616 |
histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.1 |
2.1 |
GO:0035329 |
hippo signaling(GO:0035329) |
| 0.1 |
0.5 |
GO:0031639 |
plasminogen activation(GO:0031639) |
| 0.0 |
0.1 |
GO:1904879 |
positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 |
0.2 |
GO:0070345 |
negative regulation of fat cell proliferation(GO:0070345) negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.0 |
0.5 |
GO:0098903 |
regulation of membrane repolarization during action potential(GO:0098903) |
| 0.0 |
0.6 |
GO:0060481 |
lobar bronchus epithelium development(GO:0060481) |
| 0.0 |
0.2 |
GO:0032020 |
ISG15-protein conjugation(GO:0032020) |
| 0.0 |
0.8 |
GO:0048172 |
regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 |
0.2 |
GO:0070125 |
mitochondrial translational elongation(GO:0070125) |
| 0.0 |
1.3 |
GO:0048791 |
calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 |
0.3 |
GO:1903025 |
regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.0 |
0.2 |
GO:0089711 |
L-glutamate transmembrane transport(GO:0089711) |
| 0.0 |
0.2 |
GO:0006543 |
glutamine catabolic process(GO:0006543) |
| 0.0 |
0.2 |
GO:0021508 |
floor plate formation(GO:0021508) spinal cord ventral commissure morphogenesis(GO:0021965) floor plate morphogenesis(GO:0033505) |
| 0.0 |
0.2 |
GO:0075525 |
viral translational termination-reinitiation(GO:0075525) |
| 0.0 |
1.1 |
GO:0046677 |
response to antibiotic(GO:0046677) |
| 0.0 |
0.6 |
GO:0016188 |
synaptic vesicle maturation(GO:0016188) |
| 0.0 |
0.7 |
GO:0050909 |
sensory perception of taste(GO:0050909) |
| 0.0 |
0.4 |
GO:0061302 |
smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 |
0.1 |
GO:0016332 |
establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.0 |
0.3 |
GO:0030035 |
microspike assembly(GO:0030035) |
| 0.0 |
0.2 |
GO:1902031 |
regulation of NADP metabolic process(GO:1902031) |
| 0.0 |
0.2 |
GO:0016080 |
synaptic vesicle targeting(GO:0016080) |
| 0.0 |
0.1 |
GO:0060017 |
parathyroid gland development(GO:0060017) |
| 0.0 |
0.1 |
GO:0002842 |
T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) positive regulation of T cell mediated immune response to tumor cell(GO:0002842) |
| 0.0 |
0.6 |
GO:0007614 |
short-term memory(GO:0007614) |
| 0.0 |
0.2 |
GO:0031547 |
brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.0 |
0.3 |
GO:0014842 |
regulation of skeletal muscle satellite cell proliferation(GO:0014842) |
| 0.0 |
0.7 |
GO:0070979 |
protein K11-linked ubiquitination(GO:0070979) |
| 0.0 |
0.2 |
GO:0018065 |
protein-cofactor linkage(GO:0018065) |
| 0.0 |
0.5 |
GO:0006978 |
DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) |
| 0.0 |
0.1 |
GO:0016264 |
gap junction assembly(GO:0016264) |
| 0.0 |
2.4 |
GO:2001237 |
negative regulation of extrinsic apoptotic signaling pathway(GO:2001237) |
| 0.0 |
0.1 |
GO:0048630 |
skeletal muscle tissue growth(GO:0048630) |
| 0.0 |
0.2 |
GO:2000491 |
positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 |
0.2 |
GO:0032049 |
cardiolipin biosynthetic process(GO:0032049) |
| 0.0 |
0.1 |
GO:0030854 |
positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 |
0.2 |
GO:0034498 |
early endosome to Golgi transport(GO:0034498) |
| 0.0 |
0.2 |
GO:0036158 |
outer dynein arm assembly(GO:0036158) |
| 0.0 |
0.4 |
GO:0048488 |
synaptic vesicle endocytosis(GO:0048488) |
| 0.0 |
0.5 |
GO:0043171 |
peptide catabolic process(GO:0043171) |
| 0.0 |
0.3 |
GO:0032060 |
bleb assembly(GO:0032060) |
| 0.0 |
0.2 |
GO:0048733 |
sebaceous gland development(GO:0048733) |
| 0.0 |
0.2 |
GO:0007020 |
microtubule nucleation(GO:0007020) |
| 0.0 |
0.1 |
GO:0016584 |
nucleosome positioning(GO:0016584) |
| 0.0 |
0.3 |
GO:0001829 |
trophectodermal cell differentiation(GO:0001829) |
| 0.0 |
0.2 |
GO:0044068 |
modulation by symbiont of host cellular process(GO:0044068) |
| 0.0 |
0.1 |
GO:0007602 |
phototransduction(GO:0007602) |