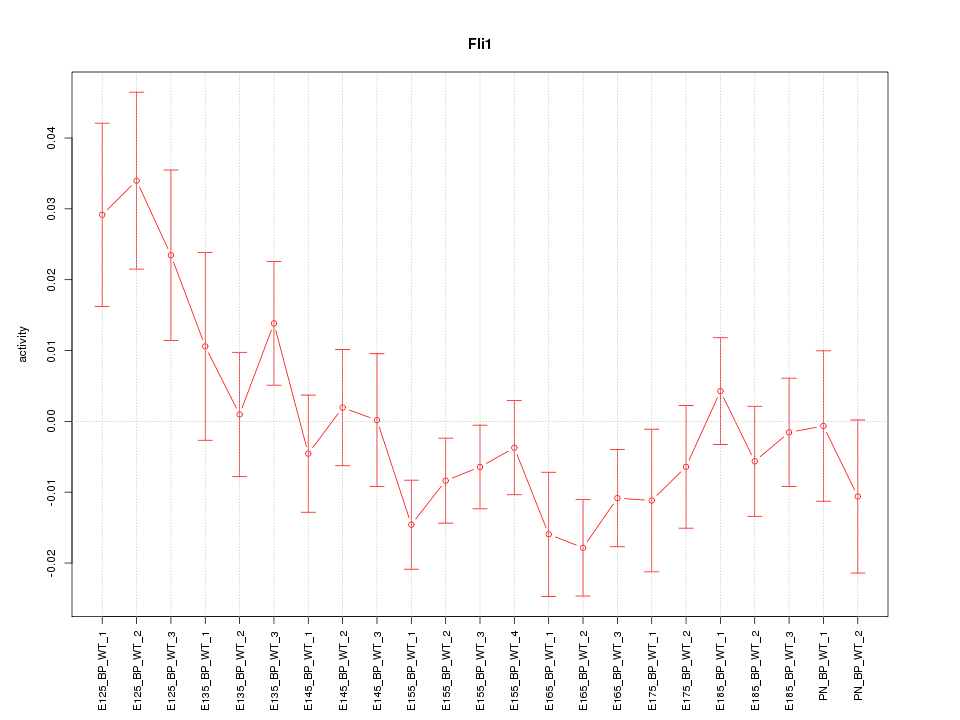
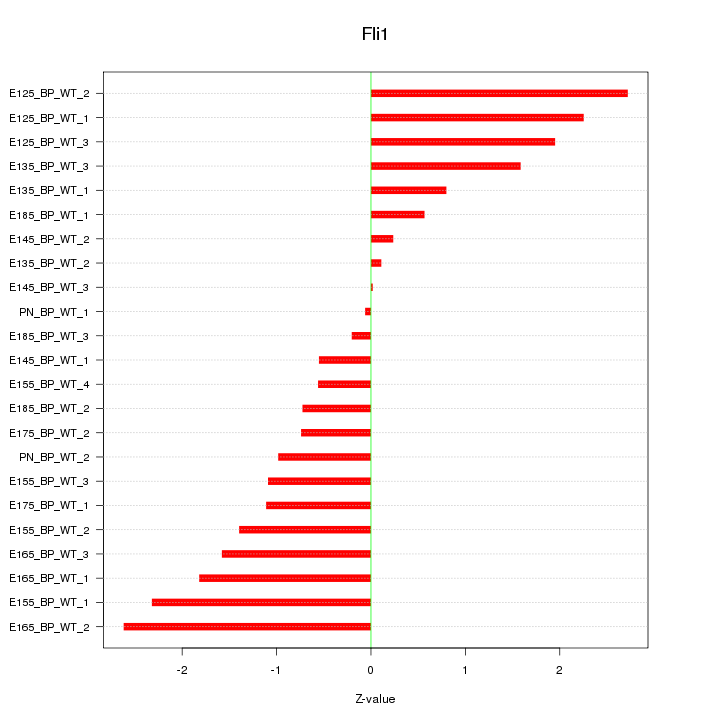
Motif ID: Fli1
Z-value: 1.398
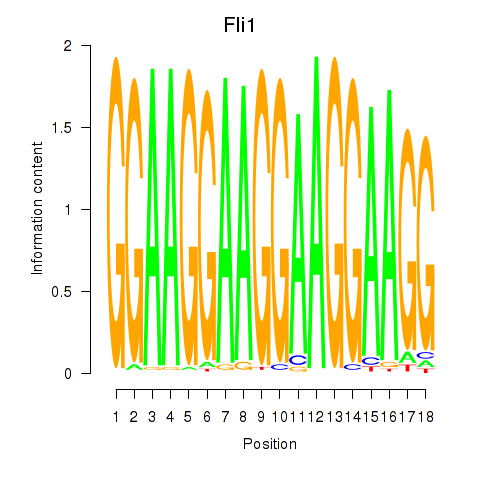
Transcription factors associated with Fli1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Fli1 | ENSMUSG00000016087.7 | Fli1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Fli1 | mm10_v2_chr9_-_32541589_32541602 | -0.50 | 1.6e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:0036118 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 1.1 | 4.4 | GO:0003360 | brainstem development(GO:0003360) |
| 1.1 | 3.3 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.7 | 2.8 | GO:0002924 | negative regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002924) |
| 0.6 | 1.9 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.6 | 4.7 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.4 | 1.3 | GO:0000087 | mitotic M phase(GO:0000087) |
| 0.3 | 1.4 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.3 | 1.3 | GO:0032901 | positive regulation of neurotrophin production(GO:0032901) |
| 0.3 | 6.4 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.3 | 1.4 | GO:0072015 | ciliary body morphogenesis(GO:0061073) glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.3 | 2.3 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.2 | 3.0 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.2 | 28.3 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.2 | 0.6 | GO:0034287 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.2 | 0.6 | GO:0048211 | Golgi vesicle docking(GO:0048211) |
| 0.2 | 0.8 | GO:0071500 | cellular response to nitrosative stress(GO:0071500) |
| 0.1 | 0.6 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.1 | 0.6 | GO:0015786 | UDP-glucose transport(GO:0015786) |
| 0.1 | 1.9 | GO:0031017 | exocrine pancreas development(GO:0031017) |
| 0.1 | 0.7 | GO:0051661 | maintenance of centrosome location(GO:0051661) regulation of microtubule motor activity(GO:2000574) |
| 0.1 | 1.5 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.1 | 1.0 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.1 | 0.6 | GO:2000774 | positive regulation of Rac protein signal transduction(GO:0035022) positive regulation of cellular senescence(GO:2000774) |
| 0.1 | 1.0 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 | 3.1 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 4.6 | GO:0033014 | porphyrin-containing compound biosynthetic process(GO:0006779) tetrapyrrole biosynthetic process(GO:0033014) |
| 0.1 | 0.6 | GO:0032261 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.1 | 0.5 | GO:1904690 | adenosine to inosine editing(GO:0006382) regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.1 | 2.1 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 0.3 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.1 | 0.5 | GO:0035087 | siRNA loading onto RISC involved in RNA interference(GO:0035087) |
| 0.1 | 2.7 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 0.2 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.1 | 3.3 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.1 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.1 | 0.3 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.1 | 0.4 | GO:0042518 | negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.1 | 0.8 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.3 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 3.2 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.3 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 1.0 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 1.5 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 3.8 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.0 | 0.6 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.1 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 1.5 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.0 | 1.0 | GO:0015991 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 | 0.2 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 9.5 | GO:0042391 | regulation of membrane potential(GO:0042391) |
| 0.0 | 0.1 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.2 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 1.6 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 0.2 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 1.1 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.6 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 3.5 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 4.9 | GO:0007283 | spermatogenesis(GO:0007283) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 28.3 | GO:0044292 | dendrite terminus(GO:0044292) |
| 1.5 | 4.4 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.7 | 5.7 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.2 | 2.5 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.2 | 4.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.2 | 4.5 | GO:0001741 | XY body(GO:0001741) |
| 0.2 | 3.1 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 0.4 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.1 | 1.0 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.1 | 0.7 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.1 | 0.3 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.1 | 1.6 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.1 | 0.2 | GO:0033193 | Lsd1/2 complex(GO:0033193) |
| 0.1 | 0.5 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.1 | 0.5 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.1 | 1.9 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 1.0 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 0.1 | 0.6 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.3 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.6 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.3 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 2.1 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 2.1 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 20.5 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 1.3 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 0.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.3 | GO:0000922 | spindle pole(GO:0000922) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 25.9 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 1.1 | 3.3 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.7 | 4.4 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.6 | 3.8 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.6 | 4.5 | GO:0051718 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.4 | 2.3 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.4 | 1.9 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.3 | 3.3 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.2 | 0.6 | GO:0019002 | GMP binding(GO:0019002) |
| 0.2 | 15.7 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.2 | 3.1 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.2 | 1.3 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.2 | 0.5 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.2 | 1.0 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.1 | 1.4 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 4.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.7 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 0.3 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.1 | 0.6 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) |
| 0.1 | 0.8 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 1.0 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.5 | GO:0019841 | retinol binding(GO:0019841) |
| 0.1 | 0.6 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 2.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 0.2 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.1 | 1.0 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.1 | 4.1 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 0.2 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.1 | 2.1 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 0.2 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.3 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 2.5 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 1.9 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 1.0 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 2.7 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 1.1 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 1.0 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.3 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.1 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.6 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.2 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.3 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.0 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.6 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 1.3 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |