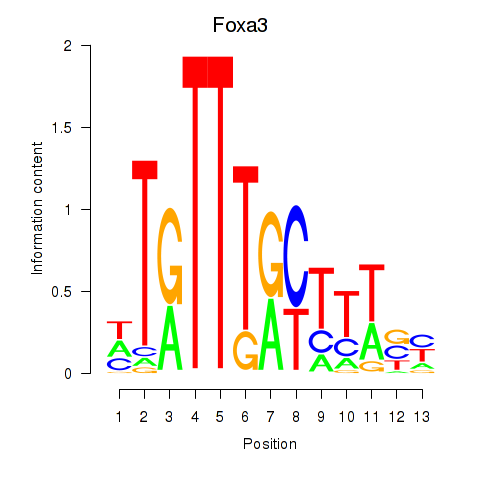
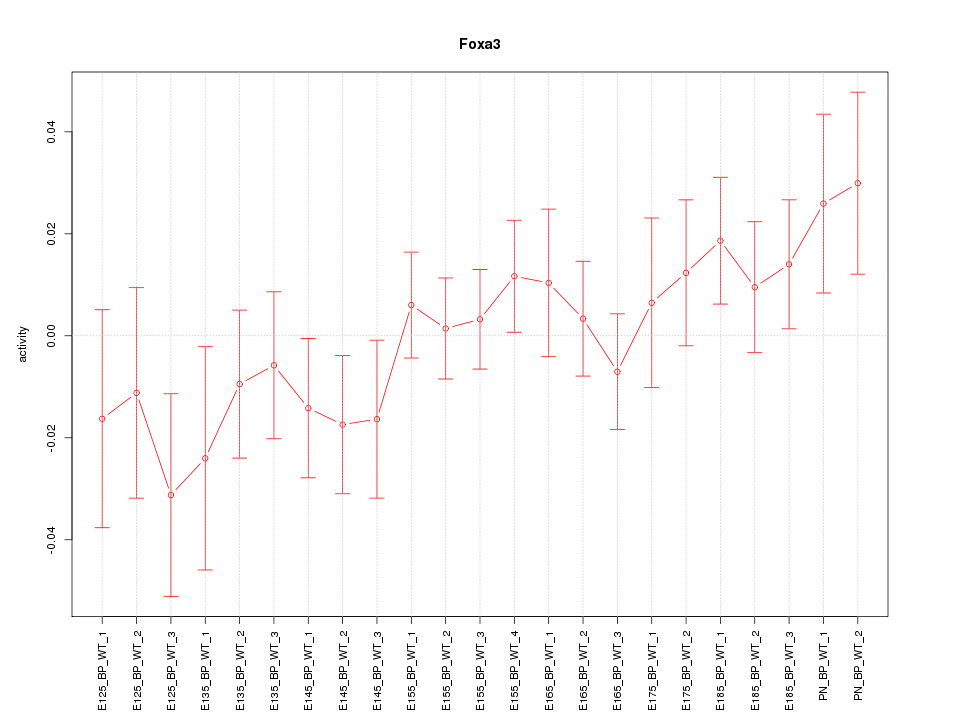
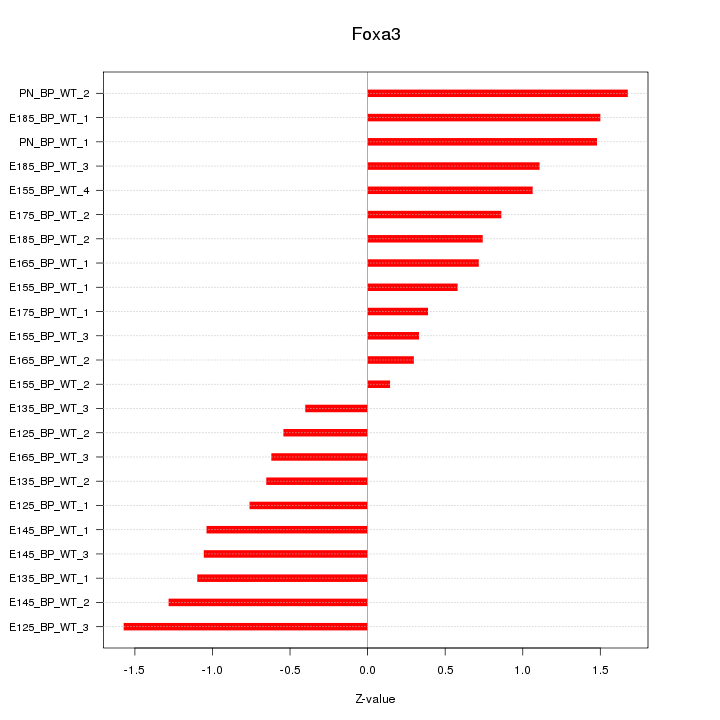
| 2.2 |
8.7 |
GO:1901894 |
regulation of calcium-transporting ATPase activity(GO:1901894) |
| 0.9 |
4.4 |
GO:2000483 |
negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.7 |
4.7 |
GO:0001887 |
selenium compound metabolic process(GO:0001887) |
| 0.7 |
2.0 |
GO:0048014 |
Tie signaling pathway(GO:0048014) |
| 0.5 |
4.2 |
GO:0031339 |
negative regulation of vesicle fusion(GO:0031339) |
| 0.5 |
3.3 |
GO:0034638 |
phosphatidylcholine catabolic process(GO:0034638) |
| 0.4 |
1.3 |
GO:0019482 |
beta-alanine metabolic process(GO:0019482) |
| 0.4 |
1.2 |
GO:1903225 |
negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.4 |
5.0 |
GO:0021684 |
cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.3 |
3.4 |
GO:0007379 |
segment specification(GO:0007379) |
| 0.3 |
1.1 |
GO:0010510 |
regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.3 |
2.1 |
GO:0097460 |
positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) ferrous iron import into cell(GO:0097460) |
| 0.3 |
0.8 |
GO:0060084 |
synaptic transmission involved in micturition(GO:0060084) |
| 0.2 |
1.0 |
GO:0060066 |
oviduct development(GO:0060066) |
| 0.2 |
0.7 |
GO:1904059 |
regulation of locomotor rhythm(GO:1904059) |
| 0.2 |
0.9 |
GO:0034285 |
response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.2 |
0.9 |
GO:0031509 |
telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.2 |
1.1 |
GO:0042636 |
negative regulation of hair cycle(GO:0042636) progesterone secretion(GO:0042701) negative regulation of hair follicle development(GO:0051799) positive regulation of ovulation(GO:0060279) |
| 0.2 |
2.0 |
GO:0097421 |
liver regeneration(GO:0097421) |
| 0.2 |
1.5 |
GO:0006488 |
dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.2 |
1.5 |
GO:0045075 |
interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.2 |
2.3 |
GO:0070842 |
aggresome assembly(GO:0070842) |
| 0.2 |
1.6 |
GO:0010820 |
positive regulation of T cell chemotaxis(GO:0010820) |
| 0.1 |
1.3 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 |
1.8 |
GO:0032695 |
negative regulation of interleukin-12 production(GO:0032695) |
| 0.1 |
0.5 |
GO:2001032 |
regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.1 |
0.7 |
GO:0001867 |
complement activation, lectin pathway(GO:0001867) |
| 0.1 |
0.6 |
GO:1904715 |
negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.1 |
0.5 |
GO:0036438 |
maintenance of lens transparency(GO:0036438) |
| 0.1 |
0.7 |
GO:0035948 |
positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of cellular ketone metabolic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072366) |
| 0.1 |
0.3 |
GO:0032929 |
negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 |
0.5 |
GO:0097368 |
establishment of Sertoli cell barrier(GO:0097368) |
| 0.1 |
0.9 |
GO:0014807 |
regulation of somitogenesis(GO:0014807) |
| 0.1 |
0.4 |
GO:0006741 |
NADP biosynthetic process(GO:0006741) |
| 0.1 |
0.2 |
GO:1901509 |
regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.1 |
0.9 |
GO:1901621 |
negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.1 |
0.2 |
GO:0043651 |
linoleic acid metabolic process(GO:0043651) |
| 0.1 |
2.1 |
GO:0035641 |
locomotory exploration behavior(GO:0035641) |
| 0.1 |
1.6 |
GO:0006883 |
cellular sodium ion homeostasis(GO:0006883) |
| 0.1 |
0.5 |
GO:0007220 |
Notch receptor processing(GO:0007220) |
| 0.1 |
0.5 |
GO:0060903 |
positive regulation of meiosis I(GO:0060903) |
| 0.1 |
0.3 |
GO:2000271 |
positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.1 |
0.3 |
GO:0051012 |
microtubule sliding(GO:0051012) negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 0.1 |
0.4 |
GO:1904217 |
regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.1 |
0.3 |
GO:1990564 |
protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.1 |
0.6 |
GO:1990440 |
positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 |
1.2 |
GO:0007205 |
protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 |
0.5 |
GO:0071763 |
nuclear membrane organization(GO:0071763) |
| 0.0 |
0.2 |
GO:0032914 |
positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.0 |
4.0 |
GO:0048663 |
neuron fate commitment(GO:0048663) |
| 0.0 |
0.8 |
GO:0030213 |
hyaluronan biosynthetic process(GO:0030213) |
| 0.0 |
0.9 |
GO:0002089 |
lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 |
0.2 |
GO:1903599 |
positive regulation of mitophagy(GO:1903599) |
| 0.0 |
0.4 |
GO:0090557 |
establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 |
0.5 |
GO:0022027 |
interkinetic nuclear migration(GO:0022027) |
| 0.0 |
3.8 |
GO:0072332 |
intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 |
0.5 |
GO:0060394 |
negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 |
0.1 |
GO:1903215 |
regulation of mRNA modification(GO:0090365) negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.0 |
0.8 |
GO:0000413 |
protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 |
0.1 |
GO:0010920 |
negative regulation of inositol phosphate biosynthetic process(GO:0010920) |
| 0.0 |
0.1 |
GO:0015803 |
branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 |
1.7 |
GO:0043268 |
positive regulation of potassium ion transport(GO:0043268) |
| 0.0 |
0.1 |
GO:0014738 |
regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 0.0 |
1.6 |
GO:0031529 |
ruffle organization(GO:0031529) |
| 0.0 |
3.6 |
GO:0051028 |
mRNA transport(GO:0051028) |
| 0.0 |
2.3 |
GO:0007601 |
visual perception(GO:0007601) |
| 0.0 |
0.5 |
GO:0008053 |
mitochondrial fusion(GO:0008053) |
| 0.0 |
0.8 |
GO:0072661 |
protein targeting to plasma membrane(GO:0072661) |
| 0.0 |
3.1 |
GO:0007160 |
cell-matrix adhesion(GO:0007160) |
| 0.0 |
0.1 |
GO:0035887 |
aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 |
0.2 |
GO:0009312 |
oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 |
0.1 |
GO:1901837 |
negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.0 |
0.5 |
GO:0006940 |
regulation of smooth muscle contraction(GO:0006940) |
| 0.0 |
0.1 |
GO:0051013 |
microtubule severing(GO:0051013) |
| 0.0 |
0.3 |
GO:0010971 |
positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.0 |
0.1 |
GO:0006297 |
nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 |
1.5 |
GO:0006813 |
potassium ion transport(GO:0006813) |
| 0.0 |
0.4 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.0 |
0.3 |
GO:0006625 |
protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |