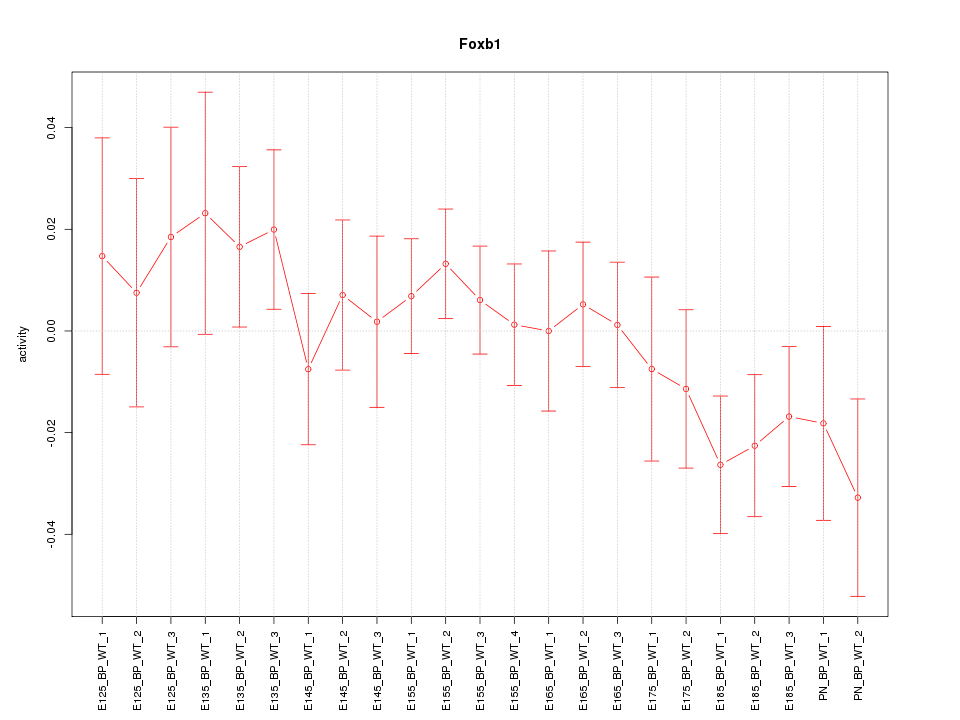
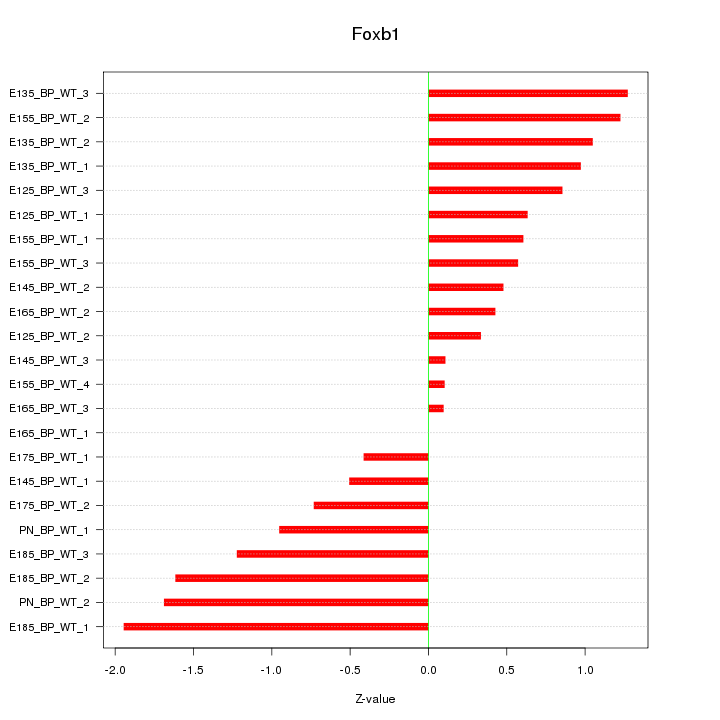
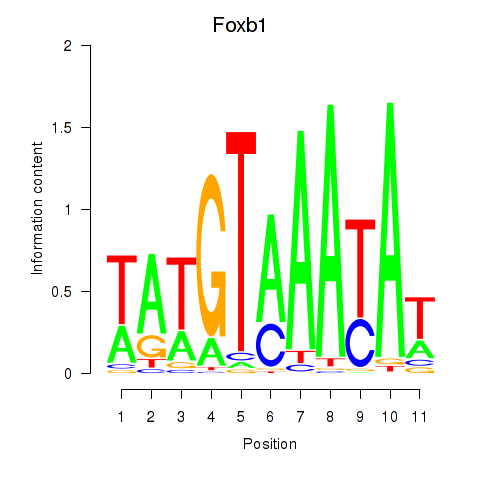
Motif ID: Foxb1
Z-value: 0.936
Transcription factors associated with Foxb1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Foxb1 | ENSMUSG00000059246.4 | Foxb1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxb1 | mm10_v2_chr9_-_69760924_69760940 | 0.22 | 3.0e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | GO:0071649 | regulation of chemokine (C-C motif) ligand 5 production(GO:0071649) positive regulation of somatostatin secretion(GO:0090274) |
| 0.4 | 1.6 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.4 | 1.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.3 | 1.0 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.3 | 1.1 | GO:0009597 | detection of virus(GO:0009597) |
| 0.3 | 3.1 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.2 | 0.9 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.2 | 2.4 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.2 | 2.0 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.2 | 1.8 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.2 | 1.9 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 0.4 | GO:0097274 | urea homeostasis(GO:0097274) |
| 0.1 | 0.4 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.1 | 1.0 | GO:0051461 | protein import into peroxisome matrix, docking(GO:0016560) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.1 | 0.8 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.1 | 2.6 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.1 | 0.8 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.1 | 1.7 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 0.3 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.1 | 0.5 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.1 | 1.7 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.1 | 0.7 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.1 | 2.3 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 0.7 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.6 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.1 | 0.7 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 0.5 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.2 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.5 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.1 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.0 | 0.4 | GO:1901898 | negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 2.8 | GO:0060349 | bone morphogenesis(GO:0060349) |
| 0.0 | 1.0 | GO:0061028 | establishment of endothelial barrier(GO:0061028) |
| 0.0 | 0.4 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 0.4 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.1 | GO:0015868 | purine nucleotide transport(GO:0015865) purine ribonucleotide transport(GO:0015868) |
| 0.0 | 0.2 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.2 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 1.6 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 0.1 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.1 | GO:0010920 | negative regulation of inositol phosphate biosynthetic process(GO:0010920) |
| 0.0 | 1.4 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.4 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 1.3 | GO:0051297 | centrosome organization(GO:0051297) |
| 0.0 | 0.1 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.0 | 3.1 | GO:0043010 | camera-type eye development(GO:0043010) |
| 0.0 | 0.3 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 1.7 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.2 | 1.0 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.2 | 1.0 | GO:0061689 | paranodal junction(GO:0033010) tricellular tight junction(GO:0061689) |
| 0.2 | 0.5 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.1 | 0.7 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.1 | 1.9 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 1.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 2.9 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.1 | 0.6 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 0.4 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.5 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 3.0 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.8 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 1.5 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.0 | 0.4 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 1.6 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.1 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.1 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 1.3 | GO:0030496 | midbody(GO:0030496) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.5 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.5 | 2.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.3 | 1.6 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.2 | 1.7 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.2 | 0.9 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.2 | 1.0 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.2 | 2.9 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.2 | 1.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.7 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 2.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 1.7 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.1 | 1.1 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 2.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.4 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.5 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 1.0 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.1 | 0.8 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 1.0 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.6 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 2.0 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.0 | 0.4 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 2.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.5 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.0 | 0.8 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 2.4 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 2.0 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 2.3 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.1 | GO:0005243 | gap junction channel activity(GO:0005243) |