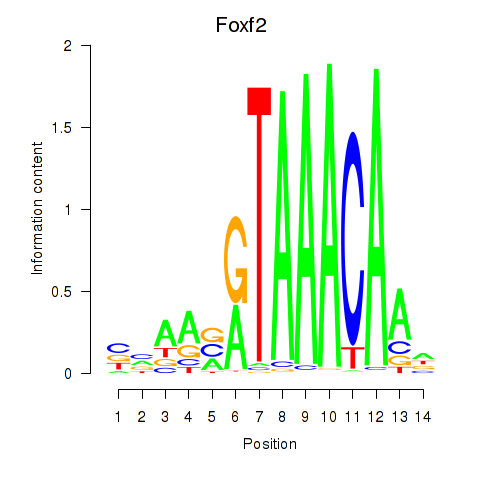
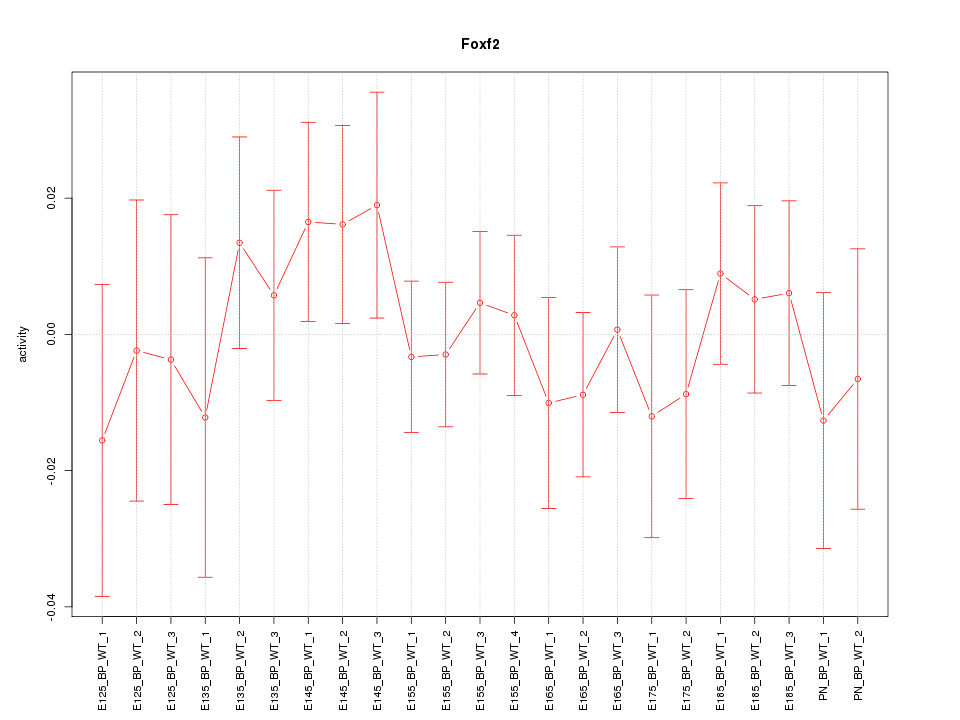
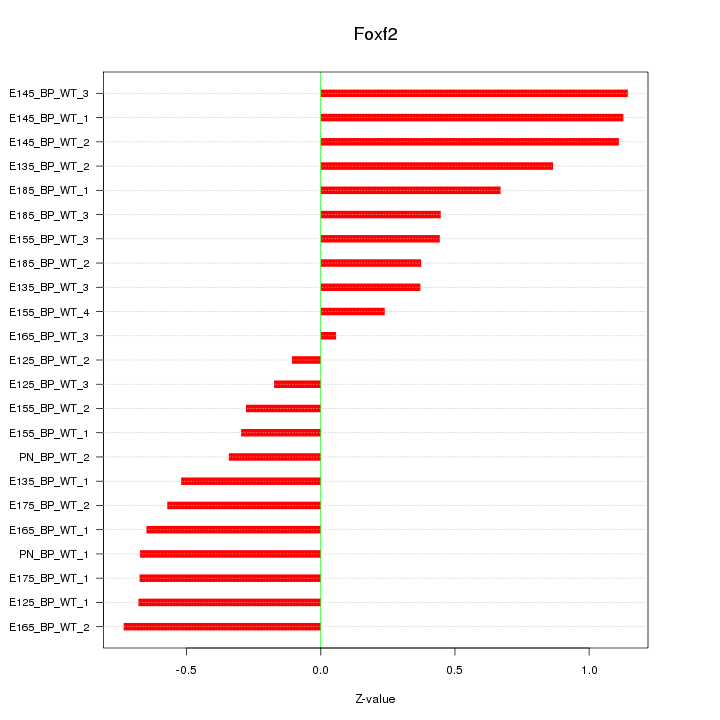
| 0.5 |
4.6 |
GO:0090232 |
positive regulation of spindle checkpoint(GO:0090232) |
| 0.3 |
1.7 |
GO:0072318 |
clathrin coat disassembly(GO:0072318) |
| 0.2 |
0.7 |
GO:0015772 |
disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.2 |
0.6 |
GO:0035582 |
sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.2 |
0.5 |
GO:0014738 |
regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 0.2 |
0.9 |
GO:0048133 |
germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.1 |
1.0 |
GO:0061002 |
negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.1 |
0.7 |
GO:0072321 |
chaperone-mediated protein transport(GO:0072321) |
| 0.1 |
1.4 |
GO:0021684 |
cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 |
0.3 |
GO:0042320 |
regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) circadian sleep/wake cycle, REM sleep(GO:0042747) positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 0.1 |
0.3 |
GO:0042271 |
susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.1 |
0.4 |
GO:0043490 |
malate-aspartate shuttle(GO:0043490) |
| 0.1 |
0.3 |
GO:1903898 |
positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.1 |
1.4 |
GO:0045792 |
negative regulation of cell size(GO:0045792) |
| 0.1 |
0.2 |
GO:0042998 |
positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.1 |
1.4 |
GO:0030213 |
hyaluronan biosynthetic process(GO:0030213) |
| 0.1 |
0.5 |
GO:0061368 |
behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.1 |
0.3 |
GO:1901740 |
negative regulation of myoblast fusion(GO:1901740) |
| 0.1 |
0.2 |
GO:0045014 |
detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) detection of glucose(GO:0051594) |
| 0.1 |
0.2 |
GO:0009597 |
detection of virus(GO:0009597) |
| 0.1 |
0.2 |
GO:0051725 |
protein de-ADP-ribosylation(GO:0051725) |
| 0.1 |
0.4 |
GO:0031642 |
negative regulation of myelination(GO:0031642) |
| 0.0 |
0.2 |
GO:0071476 |
cellular hypotonic response(GO:0071476) |
| 0.0 |
0.7 |
GO:1901739 |
regulation of myoblast fusion(GO:1901739) positive regulation of myoblast fusion(GO:1901741) |
| 0.0 |
0.4 |
GO:0032693 |
negative regulation of interleukin-10 production(GO:0032693) |
| 0.0 |
0.3 |
GO:0044027 |
hypermethylation of CpG island(GO:0044027) |
| 0.0 |
0.3 |
GO:0002755 |
MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.0 |
0.6 |
GO:0008090 |
retrograde axonal transport(GO:0008090) |
| 0.0 |
1.0 |
GO:0007608 |
sensory perception of smell(GO:0007608) |
| 0.0 |
0.2 |
GO:0007258 |
JUN phosphorylation(GO:0007258) |
| 0.0 |
0.2 |
GO:2000301 |
negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 |
0.2 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 |
0.2 |
GO:2000258 |
negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 |
0.4 |
GO:0032933 |
response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 |
0.3 |
GO:0006465 |
signal peptide processing(GO:0006465) |
| 0.0 |
0.1 |
GO:0097167 |
circadian regulation of translation(GO:0097167) |
| 0.0 |
0.2 |
GO:0045947 |
negative regulation of translational initiation(GO:0045947) |
| 0.0 |
0.4 |
GO:0071425 |
hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 |
0.0 |
GO:0002159 |
desmosome assembly(GO:0002159) |
| 0.0 |
0.1 |
GO:0010359 |
regulation of anion channel activity(GO:0010359) |
| 0.0 |
0.2 |
GO:0015937 |
coenzyme A biosynthetic process(GO:0015937) |
| 0.0 |
0.1 |
GO:1903599 |
positive regulation of mitophagy(GO:1903599) |
| 0.0 |
0.2 |
GO:2000310 |
regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |