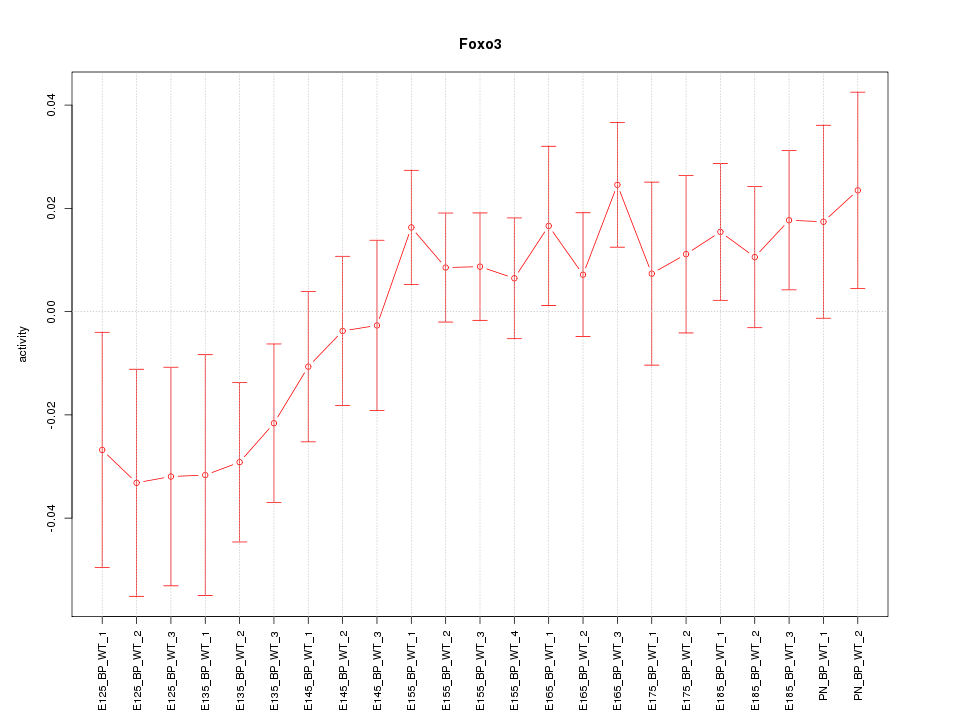
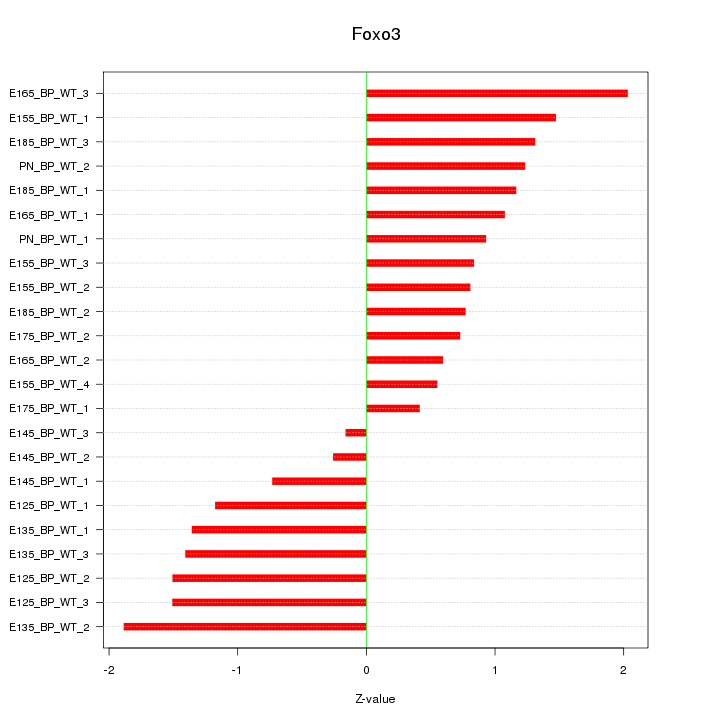
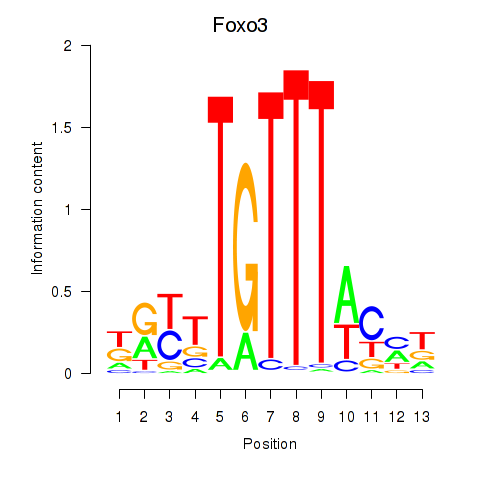
Motif ID: Foxo3
Z-value: 1.146
Transcription factors associated with Foxo3:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Foxo3 | ENSMUSG00000048756.5 | Foxo3 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxo3 | mm10_v2_chr10_-_42276744_42276763 | 0.90 | 6.4e-09 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.1 | GO:1902524 | negative regulation of interferon-alpha production(GO:0032687) interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) negative regulation of interferon-beta biosynthetic process(GO:0045358) positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 1.0 | 7.3 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.8 | 11.7 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.6 | 4.3 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.6 | 4.5 | GO:0071732 | cellular response to nitric oxide(GO:0071732) |
| 0.6 | 1.7 | GO:0003278 | apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.6 | 5.6 | GO:0007379 | segment specification(GO:0007379) |
| 0.4 | 0.9 | GO:0002741 | positive regulation of cytokine secretion involved in immune response(GO:0002741) |
| 0.4 | 2.6 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.4 | 1.8 | GO:1903726 | negative regulation of phospholipid metabolic process(GO:1903726) |
| 0.3 | 2.7 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.3 | 1.8 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.3 | 0.9 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.3 | 0.9 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.3 | 6.9 | GO:0010574 | regulation of vascular endothelial growth factor production(GO:0010574) |
| 0.2 | 1.2 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.2 | 1.2 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.2 | 0.7 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.1 | 1.9 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.1 | 2.3 | GO:0042182 | ketone catabolic process(GO:0042182) |
| 0.1 | 0.4 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.1 | 0.7 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.1 | 1.3 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 1.1 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.1 | 1.0 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.1 | 0.7 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.1 | 1.1 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.1 | 1.1 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 0.3 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.1 | 1.7 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 0.4 | GO:0032929 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 | 0.8 | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) |
| 0.0 | 0.7 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.8 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.0 | 1.4 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 | 0.6 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.0 | 0.2 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 0.0 | 2.1 | GO:0050729 | positive regulation of inflammatory response(GO:0050729) |
| 0.0 | 0.1 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.0 | 1.1 | GO:0009395 | phospholipid catabolic process(GO:0009395) |
| 0.0 | 0.1 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.2 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 1.5 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.0 | 4.7 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 0.4 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.1 | GO:0045630 | positive regulation of T-helper 2 cell differentiation(GO:0045630) |
| 0.0 | 1.0 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 2.9 | GO:0042060 | wound healing(GO:0042060) |
| 0.0 | 1.7 | GO:0008360 | regulation of cell shape(GO:0008360) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.3 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.3 | 0.8 | GO:0031251 | PAN complex(GO:0031251) |
| 0.2 | 1.3 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 4.5 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 5.6 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 0.4 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 1.1 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 2.6 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 1.0 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 1.0 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.2 | GO:0005712 | chiasma(GO:0005712) |
| 0.0 | 2.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.4 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 1.8 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 1.0 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.1 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 4.8 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 1.2 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 1.7 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 1.1 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.3 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 1.1 | 4.3 | GO:0031721 | haptoglobin binding(GO:0031720) hemoglobin alpha binding(GO:0031721) |
| 0.9 | 3.6 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.6 | 1.8 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.5 | 2.6 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.5 | 1.5 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.4 | 1.1 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.3 | 1.2 | GO:0038049 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.2 | 0.9 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.2 | 1.0 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 1.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.2 | 1.8 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.2 | 1.0 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.2 | 4.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.7 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.1 | 1.7 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 0.4 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 1.1 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 1.7 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) |
| 0.0 | 0.2 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 2.1 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.8 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 3.3 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.7 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 1.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.4 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 13.0 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.7 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 2.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.5 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 1.0 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.9 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 1.7 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 1.2 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 2.7 | GO:0019902 | phosphatase binding(GO:0019902) |