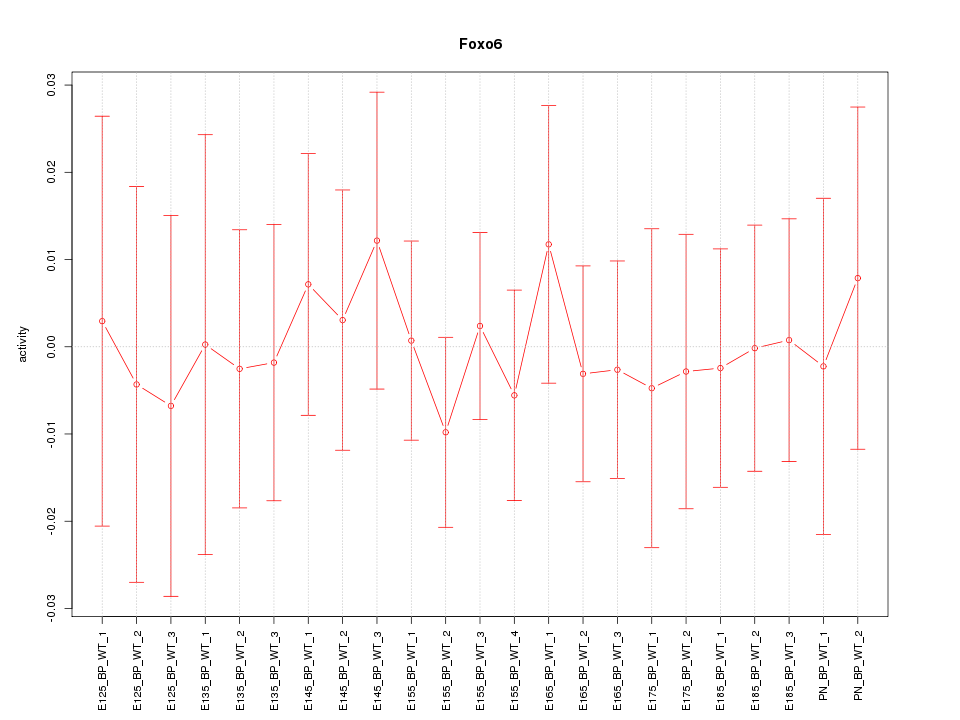
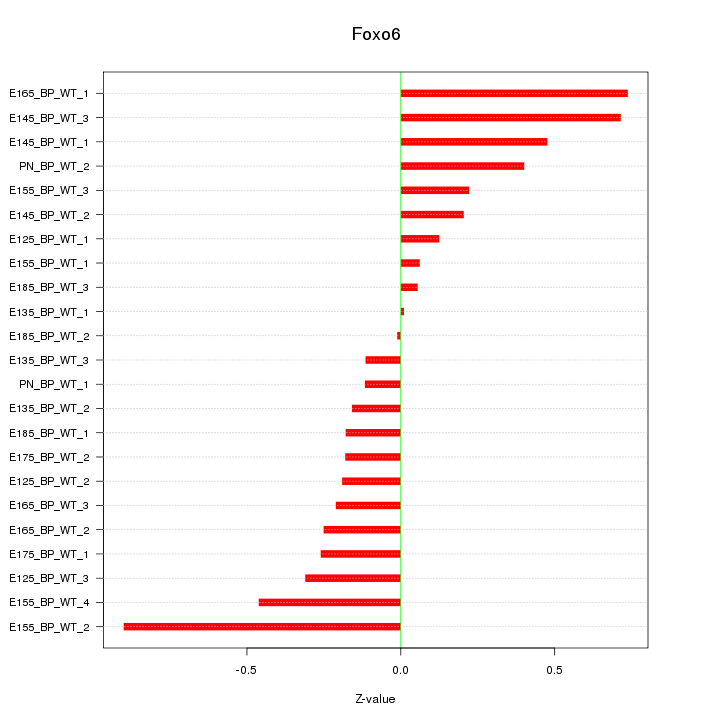
Motif ID: Foxo6
Z-value: 0.362
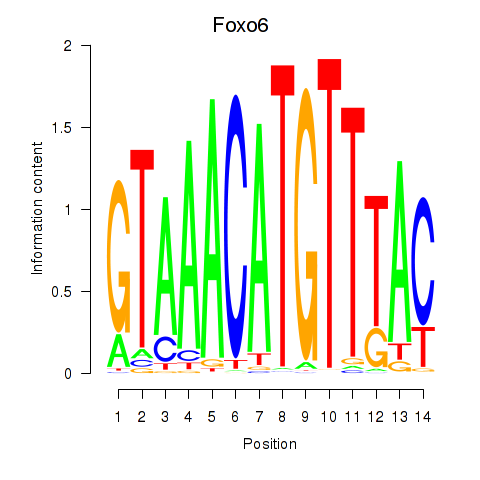
Transcription factors associated with Foxo6:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Foxo6 | ENSMUSG00000052135.8 | Foxo6 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxo6 | mm10_v2_chr4_-_120287349_120287349 | 0.20 | 3.5e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 | 0.2 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.1 | 0.6 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.2 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 0.2 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.0 | 0.3 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 0.1 | GO:0007521 | muscle cell fate determination(GO:0007521) mammary placode formation(GO:0060596) |
| 0.0 | 0.4 | GO:1901898 | negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.2 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.2 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.1 | GO:0002586 | positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) detection of peptidoglycan(GO:0032499) |
| 0.0 | 0.1 | GO:2001187 | positive regulation of CD8-positive, alpha-beta T cell activation(GO:2001187) |
| 0.0 | 0.1 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.0 | 0.1 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.0 | 0.1 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.0 | 0.1 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.0 | 0.0 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.0 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.1 | GO:0097421 | liver regeneration(GO:0097421) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.2 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.4 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.4 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.0 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.6 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.1 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 0.4 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.1 | 0.6 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.2 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.0 | 0.2 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.4 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.3 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.1 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.2 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.0 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.0 | 0.1 | GO:0070891 | peptidoglycan binding(GO:0042834) lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.2 | GO:1990459 | transferrin receptor binding(GO:1990459) |