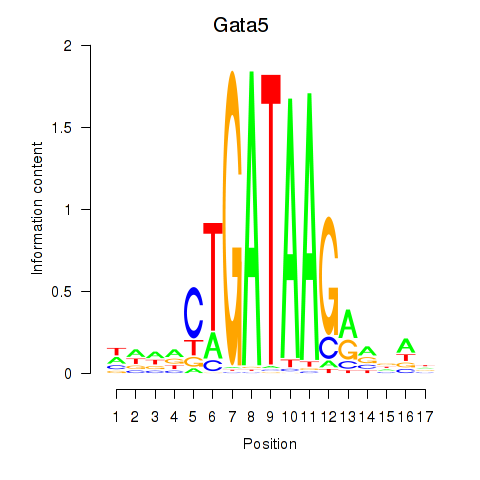
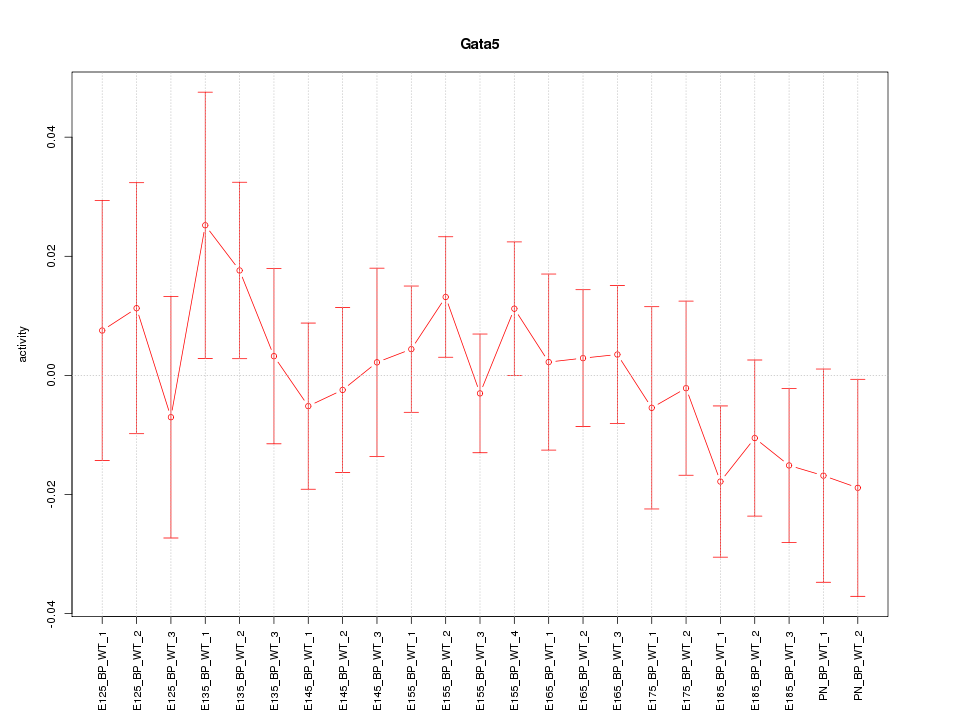
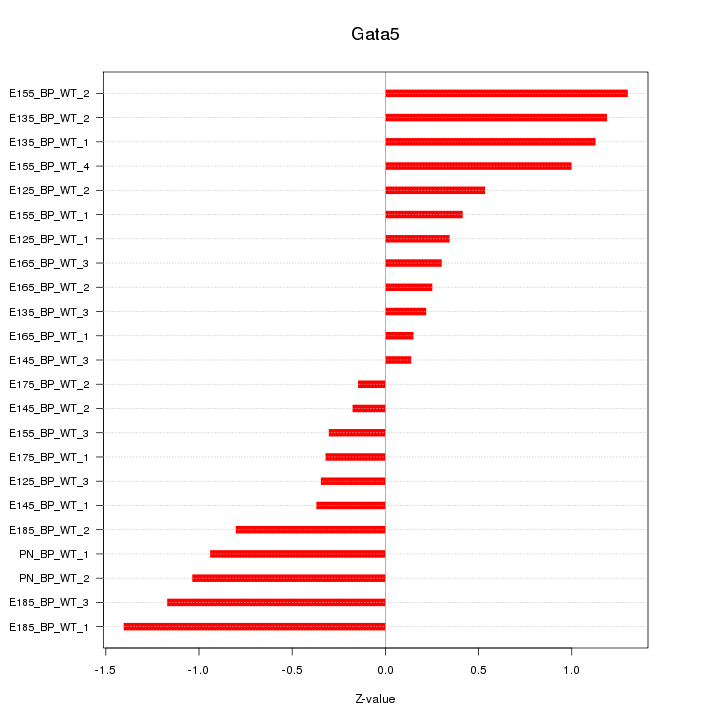
| 2.5 |
12.6 |
GO:0015671 |
oxygen transport(GO:0015671) |
| 0.5 |
2.6 |
GO:1903215 |
negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.3 |
1.6 |
GO:0060838 |
lymphatic endothelial cell fate commitment(GO:0060838) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.3 |
0.9 |
GO:0001830 |
trophectodermal cell fate commitment(GO:0001830) |
| 0.2 |
1.1 |
GO:0061343 |
cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.2 |
0.5 |
GO:0006436 |
tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 |
0.3 |
GO:1902605 |
heterotrimeric G-protein complex assembly(GO:1902605) |
| 0.1 |
0.9 |
GO:0061368 |
behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.1 |
0.3 |
GO:0060023 |
soft palate development(GO:0060023) |
| 0.1 |
2.2 |
GO:0045663 |
positive regulation of myoblast differentiation(GO:0045663) |
| 0.1 |
0.2 |
GO:0019401 |
alditol biosynthetic process(GO:0019401) |
| 0.1 |
2.7 |
GO:0003301 |
physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 |
0.5 |
GO:0006071 |
glycerol metabolic process(GO:0006071) |
| 0.1 |
0.3 |
GO:0072592 |
regulation of integrin biosynthetic process(GO:0045113) oxygen metabolic process(GO:0072592) |
| 0.1 |
0.4 |
GO:0032229 |
positive regulation of gamma-aminobutyric acid secretion(GO:0014054) negative regulation of synaptic transmission, GABAergic(GO:0032229) |
| 0.1 |
0.5 |
GO:0002862 |
negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.1 |
0.2 |
GO:0019805 |
quinolinate biosynthetic process(GO:0019805) |
| 0.0 |
0.1 |
GO:2000616 |
negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 0.0 |
0.5 |
GO:0046543 |
development of secondary female sexual characteristics(GO:0046543) |
| 0.0 |
0.3 |
GO:0042866 |
pyruvate biosynthetic process(GO:0042866) |
| 0.0 |
0.2 |
GO:0016068 |
regulation of type I hypersensitivity(GO:0001810) positive regulation of type I hypersensitivity(GO:0001812) type I hypersensitivity(GO:0016068) |
| 0.0 |
0.5 |
GO:0032693 |
negative regulation of interleukin-10 production(GO:0032693) |
| 0.0 |
0.7 |
GO:0034390 |
smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.0 |
0.4 |
GO:0046485 |
ether lipid metabolic process(GO:0046485) |
| 0.0 |
0.4 |
GO:0090527 |
actin filament reorganization(GO:0090527) |
| 0.0 |
0.2 |
GO:0045657 |
positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 |
0.5 |
GO:0009312 |
oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 |
0.3 |
GO:0034242 |
negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.0 |
0.3 |
GO:0031468 |
nuclear envelope reassembly(GO:0031468) |
| 0.0 |
0.2 |
GO:0030277 |
maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 |
0.5 |
GO:2000114 |
regulation of establishment of cell polarity(GO:2000114) |
| 0.0 |
0.2 |
GO:0043353 |
enucleate erythrocyte differentiation(GO:0043353) hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.0 |
0.1 |
GO:2000620 |
positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.0 |
2.1 |
GO:0051965 |
positive regulation of synapse assembly(GO:0051965) |
| 0.0 |
0.1 |
GO:0046501 |
protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 |
0.1 |
GO:0035026 |
leading edge cell differentiation(GO:0035026) |
| 0.0 |
0.2 |
GO:0035970 |
peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 |
0.1 |
GO:0046013 |
regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 |
2.1 |
GO:0030177 |
positive regulation of Wnt signaling pathway(GO:0030177) |
| 0.0 |
0.2 |
GO:0044406 |
adhesion of symbiont to host(GO:0044406) |
| 0.0 |
0.4 |
GO:1902043 |
positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 |
0.7 |
GO:0045773 |
positive regulation of axon extension(GO:0045773) |
| 0.0 |
0.3 |
GO:0015813 |
L-glutamate transport(GO:0015813) |